Search Count: 43
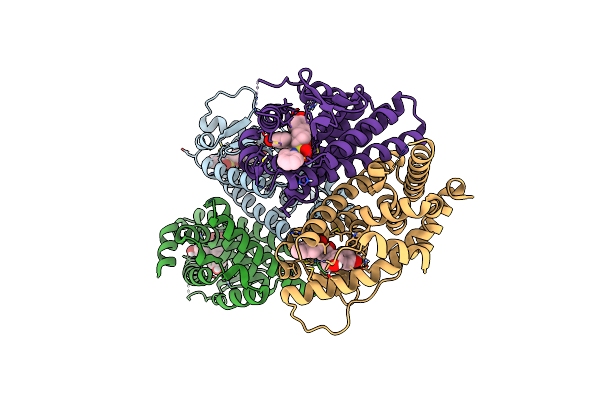 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-411
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHV |
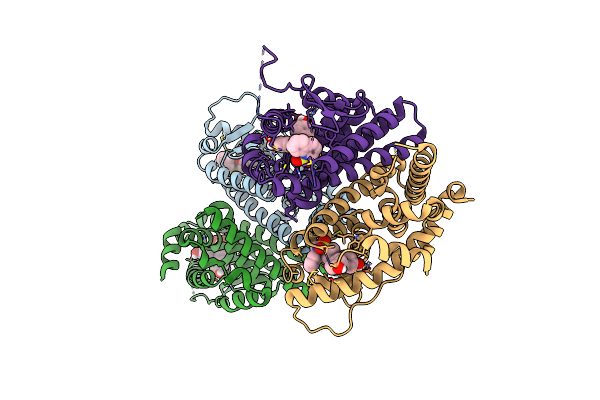 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-410
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHU |
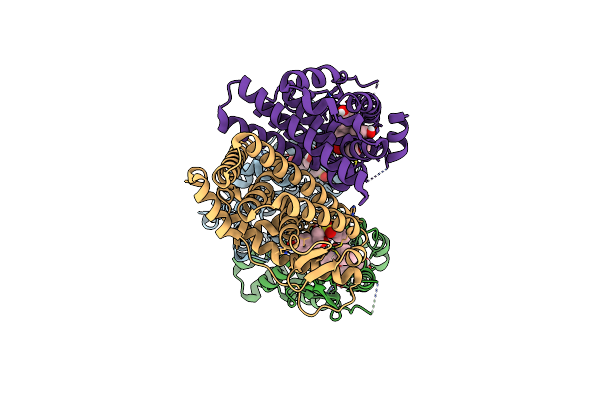 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-400
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHO |
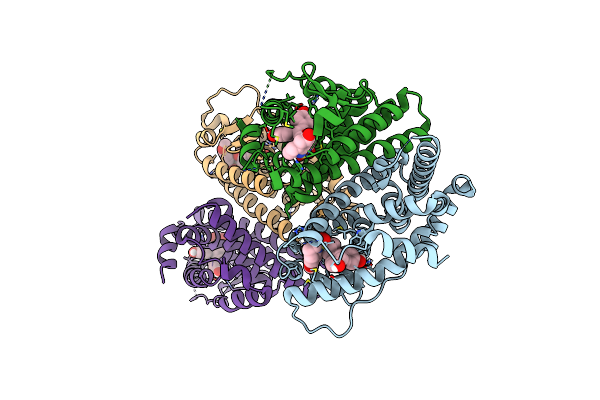 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-409
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHS |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-403
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHW |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-406
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHX, NI |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-1154
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: OBT |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With K-402
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2024-06-12 Classification: NUCLEAR PROTEIN Ligands: A1AHY, A1AHZ |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With Dmeri-19
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2021-09-22 Classification: TRANSCRIPTION/TRANSCRIPTION INHIBITOR Ligands: 7AI, CL |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With Dmeri-30
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2021-09-22 Classification: TRANSCRIPTION/TRANSCRIPTION Inhibitor Ligands: 73I |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With Dmeri-16
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2021-09-22 Classification: TRANSCRIPTION/TRANSCRIPTION INHIBITOR Ligands: 7EI |
 |
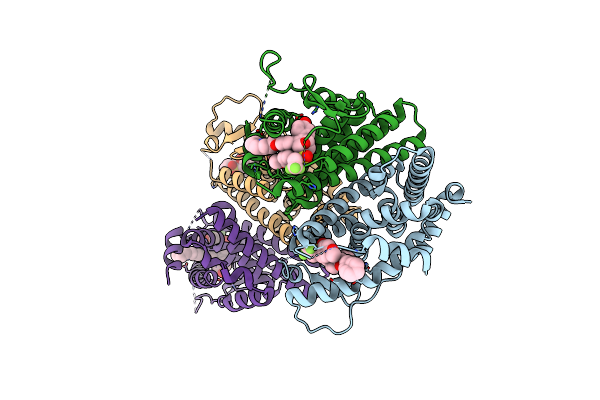
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With Dmeri-20
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2021-09-08 Classification: TRANSCRIPTION/TRANSCRIPTION INHIBITOR Ligands: L84, CL |
 |
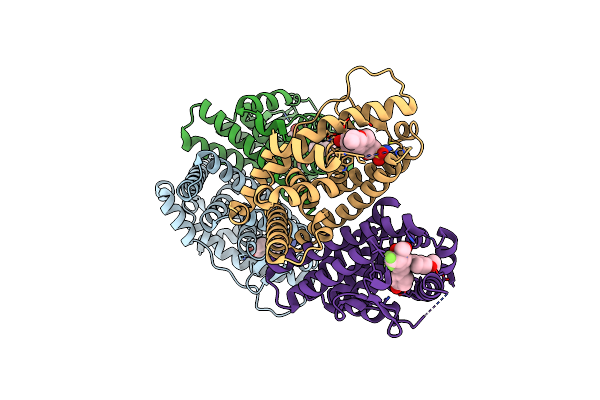
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With Dmeri-30
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2021-09-08 Classification: TRANSCRIPTION/TRANSCRIPTION INHIBITOR Ligands: 77I |
 |
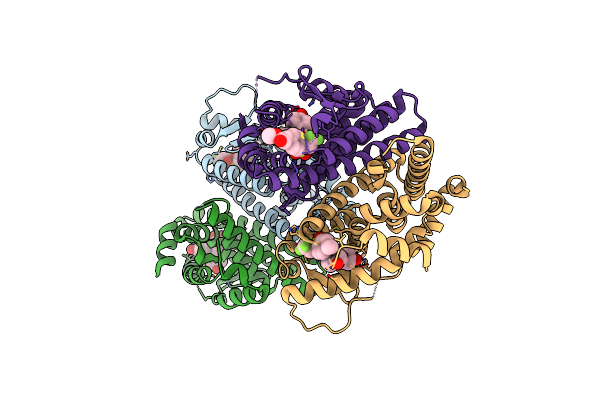
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With Dmeri-18
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2021-09-08 Classification: TRANSCRIPTION/TRANSCRIPTION INHIBITOR Ligands: 7I9 |
 |
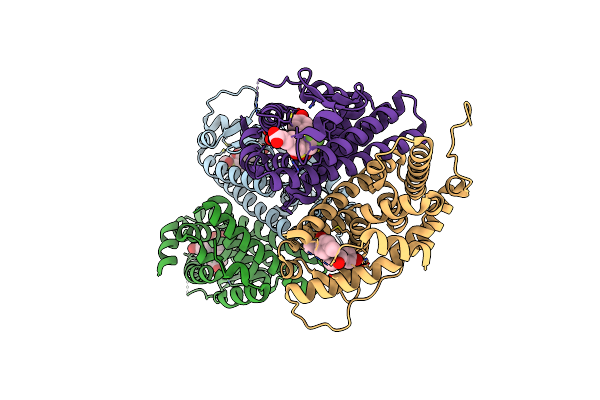
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With Dmeri-21
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2021-09-08 Classification: TRANSCRIPTION/TRANSCRIPTION INHIBITOR Ligands: 7Q5 |
 |
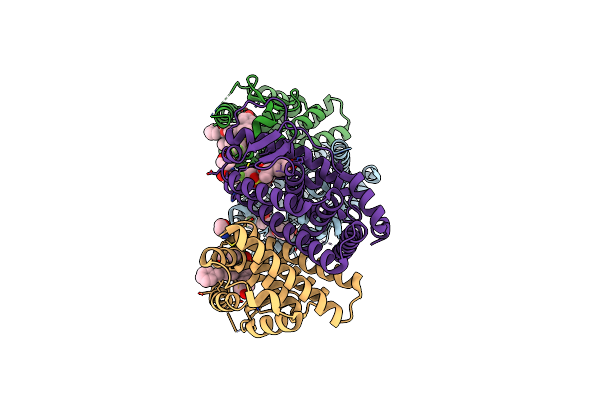
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With Dmeri-23
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2021-09-08 Classification: TRANSCRIPTION/TRANSCRIPTION INHIBITOR Ligands: 7I5 |
 |
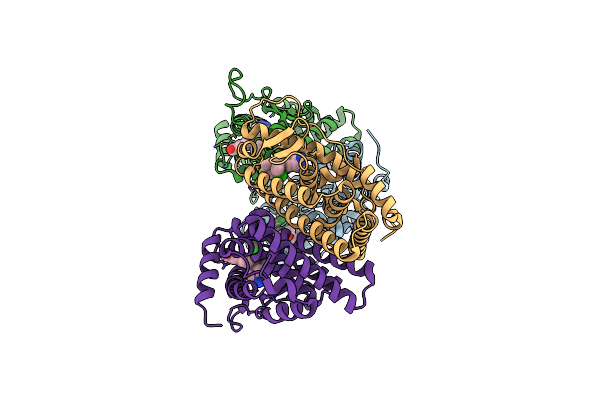
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With Dmeri-29
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2021-09-08 Classification: TRANSCRIPTION/TRANSCRIPTION INHIBITOR Ligands: 7OR, TYR, SER, CYS |
 |
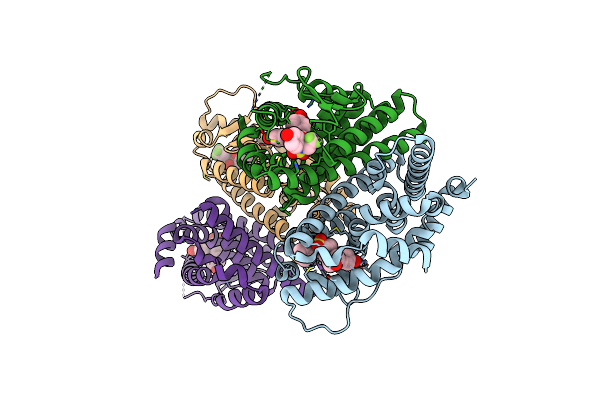
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With Dmeri-8
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2021-09-08 Classification: TRANSCRIPTION/TRANSCRIPTION INHIBITOR Ligands: 7I0, CL |
 |
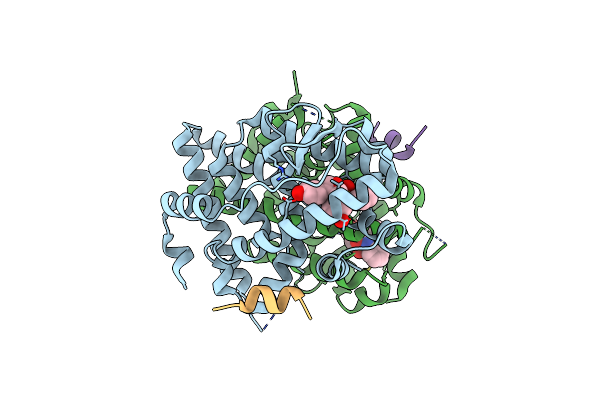
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (L372S, L536S) In Complex With Dmeri-25
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-09-08 Classification: TRANSCRIPTION/TRANSCRIPTION INHIBITOR Ligands: 7OI |
 |
Crystal Structure Of The Er-Alpha Ligand-Binding Domain (Y537S) In Complex With Oxabicyclic Heptene Sulfonamide (Obhs-N)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2016-11-16 Classification: TRANSCRIPTION Ligands: OB1 |

