Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Aeromonas sobria
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2009-06-02 Classification: HYDROLASE Ligands: CA |
 |
Organism: Thermococcus litoralis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2008-06-17 Classification: TRANSFERASE Ligands: ZN, PMP, IMD |
 |
Organism: Clostridium perfringens, Oryctolagus cuniculus
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2008-05-13 Classification: TOXIN/STRUCTURAL PROTEIN Ligands: TAD, CA, ATP, LAR |
 |
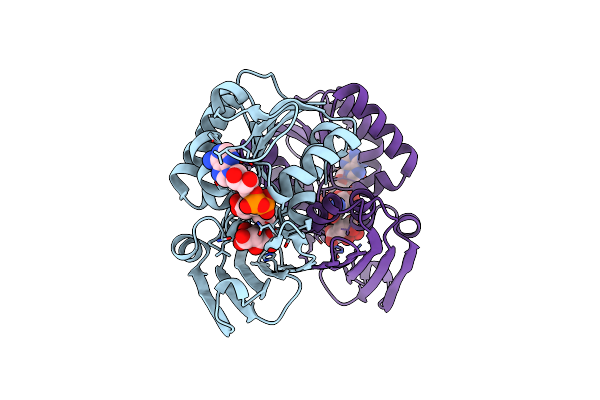
Crystal Structure Of L-Aspartate Oxidase From Hyperthermophilic Archaeon Sulfolobus Tokodaii
Organism: Sulfolobus tokodaii
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2008-01-01 Classification: OXIDOREDUCTASE Ligands: CL, FAD |
 |
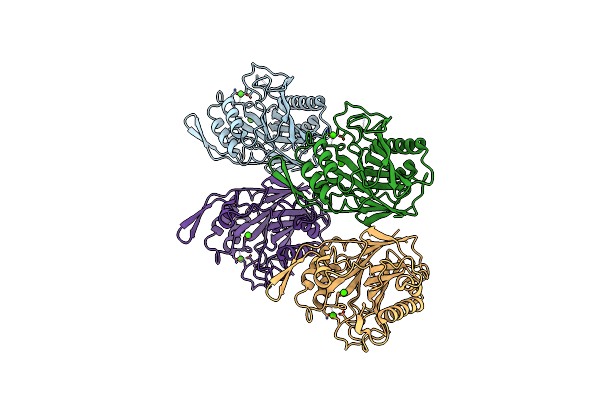
Crystal Structure Of L-Aspartate Dehydrogenase From Hyperthermophilic Archaeon Archaeoglobus Fulgidus
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2006-12-26 Classification: OXIDOREDUCTASE Ligands: CIT, NAD |
 |
Crystal Structure Of Sphingomyelinase From Bacillus Cereus With Calcium Ion
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2006-05-02 Classification: HYDROLASE Ligands: CA |
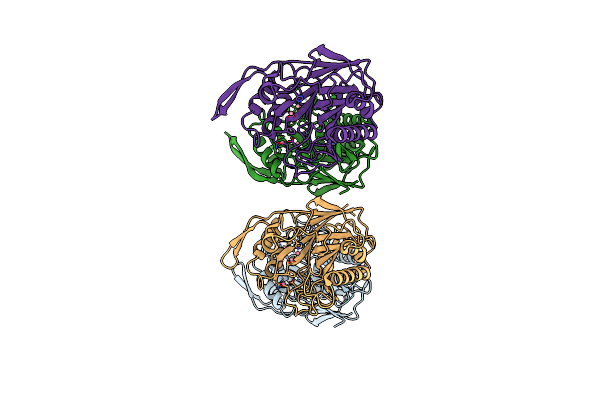 |
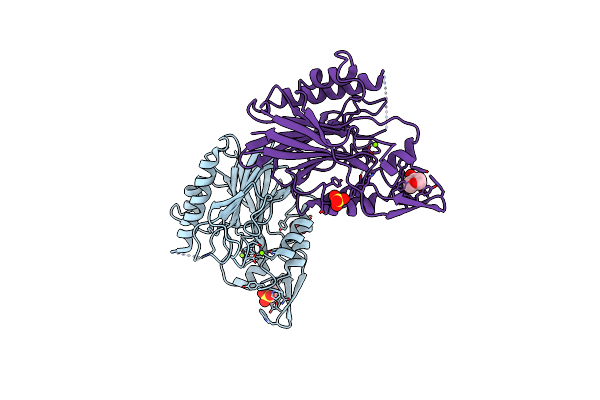
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-05-02 Classification: HYDROLASE Ligands: CO |
 |
Crystal Structure Of Sphingomyelinase From Bacillus Cereus With Magnesium Ion
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-05-02 Classification: HYDROLASE Ligands: MG, SO4, MES |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2005-07-26 Classification: OXIDOREDUCTASE Ligands: FE, SO4, FMN, ATP, CXS, CL, FAD |
 |
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2005-06-07 Classification: BIOSYNTHETIC PROTEIN Ligands: MLT |
 |
Crystal Structure Of Hypothetical Protein [St1625P] From Hyperthermophilic Archaeon Sulfolobus Tokodaii
Organism: Sulfolobus tokodaii
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2005-02-15 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |
 |
Organism: Pyrobaculum islandicum
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2004-12-14 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Unique Tetrameric Structure Of Deoxyribose Phosphate Aldolase From Aeropyrum Pernix
Organism: Aeropyrum pernix
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2003-03-25 Classification: LYASE |
 |
Crystal Structure Of The Enzymatic Componet Of Iota-Toxin From Clostridium Perfringens With Nadh
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2003-01-14 Classification: TOXIN Ligands: NAI |
 |
Crystal Structure Of The Enzymatic Componet Of Iota-Toxin From Clostridium Perfringens With Nadph
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2003-01-14 Classification: TOXIN Ligands: NDP |
 |
Crystal Structure Of Adp-Dependent Glucokinase From A Pyrococcus Horikoshii
Organism: Pyrococcus horikoshii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2002-12-30 Classification: TRANSFERASE |
 |
Crystal Structure Analysis Of Papain With Clik148(Cathepsin L Specific Inhibitor)
Organism: Carica papaya
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2000-08-30 Classification: HYDROLASE Ligands: C48 |
 |
Crystal Structure Of Cathepsin B Inhibited With Ca030 At 2.1 Angstroms Resolution: A Basis For The Design Of Specific Epoxysuccinyl Inhibitors
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1996-04-03 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: EP0 |
 |
The Refined 2.15 Angstroms X-Ray Crystal Structure Of Human Liver Cathepsin B: The Structural Basis For Its Specificity
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 1995-01-26 Classification: THIOL PROTEASE |