Search Count: 23
 |
Organism: Kutzneria albida dsm 43870
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: LIGASE Ligands: MG, AMP |
 |
Organism: Kutzneria albida dsm 43870
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: LIGASE |
 |
Crosslinked Crystal Structure Of Type Ii Fatty Acid Synthase Ketosynthase, Fabb, And C16:1-Crypto Acyl Carrier Protein, Acpp
Organism: Escherichia coli (strain k12), Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-11-23 Classification: TRANSFERASE |
 |
Crosslinked Crystal Structure Of Type Ii Fatty Acid Synthase Ketosynthase, Fabb, And C14-Crypto Acyl Carrier Protein, Acpp
Organism: Escherichia coli k-12, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-08-03 Classification: TRANSFERASE |
 |
Organism: Streptomyces sp. ri18
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-12-01 Classification: TRANSFERASE Ligands: EPE, GOL, LYS, EDO |
 |
Geranyl Pyrophosphate C6-Methyltransferase Beza Binding With S-Adenosylhomocysteine
Organism: Streptomyces sp. ri-18-2
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2021-12-01 Classification: TRANSFERASE Ligands: EPE, SAH |
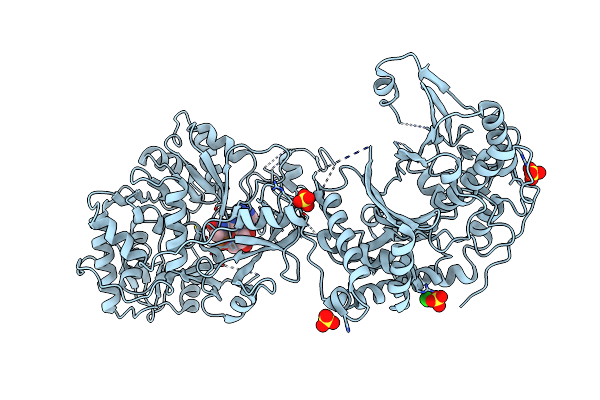 |
Crystal Structure Of Nonribosomal Peptide Synthetases (Nrps), Fmoa3 (S1046A)-Alpha-Methyl-L-Serine-Amp Bound Form
Organism: Streptomyces sp. sp080513ge-23
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2021-03-10 Classification: BIOSYNTHETIC PROTEIN Ligands: AMP, EW6, SO4, CL |
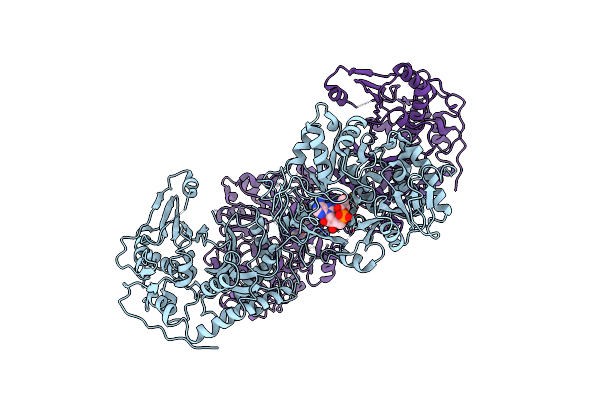 |
Crystal Structure Of Nonribosomal Peptide Synthetases (Nrps), Fmoa3 (S1046A)-Alpha-Methyl-L-Serine-Amp Bound Form
Organism: Streptomyces sp. sp080513ge-23
Method: X-RAY DIFFRACTION Resolution:4.10 Å Release Date: 2021-03-10 Classification: BIOSYNTHETIC PROTEIN Ligands: AMP, EW6 |
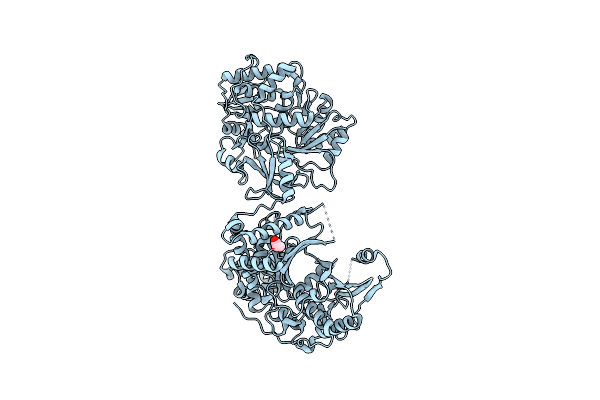 |
Crystal Structure Of Nonribosomal Peptide Synthetases (Nrps), Fmoa3 (S1046A)
Organism: Streptomyces sp. sp080513ge-23
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2021-02-24 Classification: BIOSYNTHETIC PROTEIN Ligands: AKR |
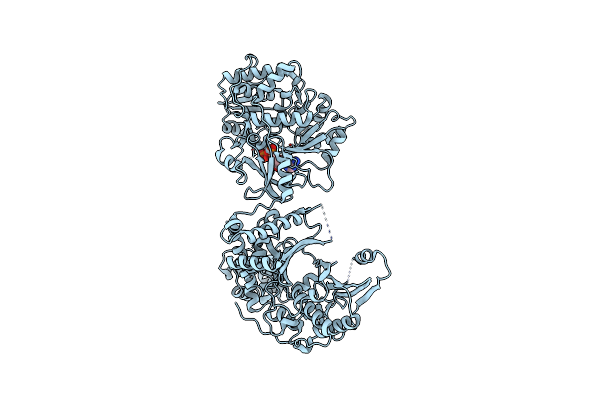 |
Crystal Structure Of Nonribosomal Peptide Synthetases (Nrps), Fmoa3 (S1046A)-Amppnp Bound Form
Organism: Streptomyces sp. sp080513ge-23
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2021-02-24 Classification: BIOSYNTHETIC PROTEIN Ligands: ANP |
 |
Organism: Streptomyces sp. msc090213je08
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-05-06 Classification: TRANSFERASE Ligands: PO4, EDO, ACT |
 |
Organism: Streptomyces sp. msc090213je08
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2020-05-06 Classification: TRANSFERASE Ligands: PPI, ACT, EDO, PO4 |
 |
Organism: Streptomyces sp. msc090213je08
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2020-05-06 Classification: TRANSFERASE Ligands: DYF |
 |
Organism: Streptomyces cremeus
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2018-03-14 Classification: LYASE |
 |
Organism: Streptomyces cremeus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-03-14 Classification: LYASE Ligands: FUM |
 |
Organism: Streptomyces sp. dsm 40835
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2017-10-04 Classification: OXIDOREDUCTASE |
 |
Organism: Streptomyces sp. dsm 40835
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2017-10-04 Classification: OXIDOREDUCTASE Ligands: CO, AKG, LEU, NA |
 |
Organism: Streptomyces sp. dsm 40835
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2017-10-04 Classification: OXIDOREDUCTASE Ligands: MN, SIN, HL5 |
 |
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2013-04-24 Classification: TRANSFERASE |
 |
Organism: Azotobacter vinelandii
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2013-04-24 Classification: TRANSFERASE |

