Search Count: 36
 |
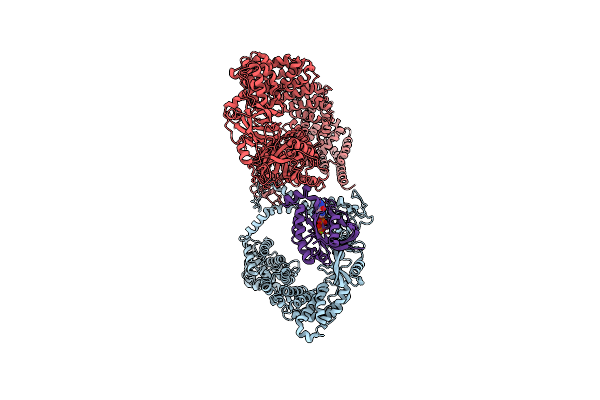
Cryo-Em Structure Of Tmprss2 In Complex With Fab Fragments Of 752 Mab And 2228 Mab
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: MEMBRANE PROTEIN |
 |
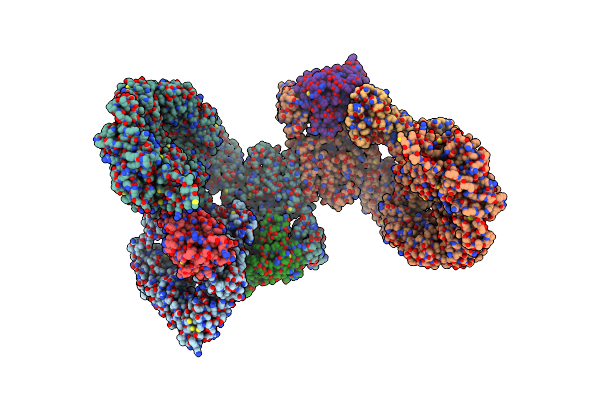
Cryo-Em Structure Of The Rhog/Dock5/Elmo1/Rac1 Complex: Rhog/Dock5/Elmo1 Focused Map
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: SIGNALING PROTEIN Ligands: MG, GTP |
 |
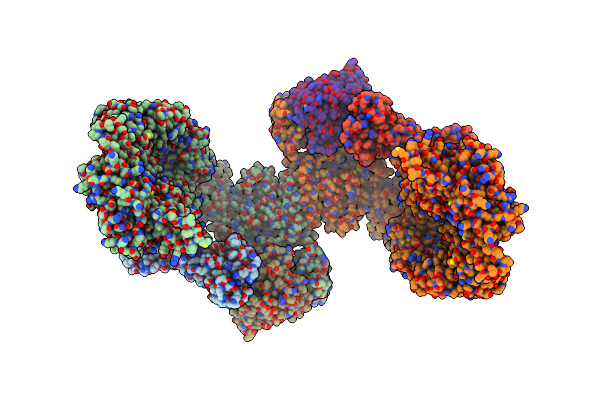
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: SIGNALING PROTEIN Ligands: MG, GTP |
 |
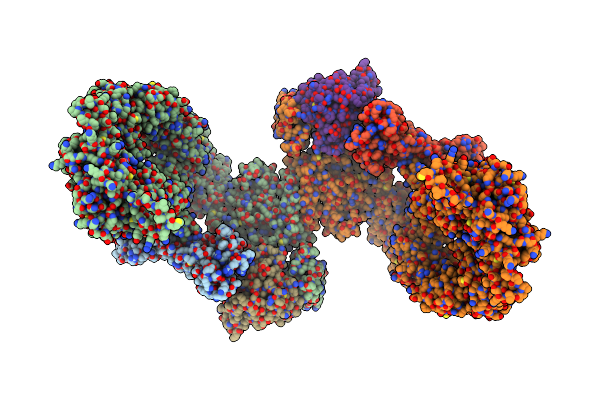
Structure Of Dock5/Elmo1/Rac1 Core (Rhog/Dock5/Elmo1/Rac1 Dataset, Class 1)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: SIGNALING PROTEIN |
 |
Structure Of Dock5/Elmo1/Rac1 Core (Rhog/Dock5/Elmo1/Rac1 Dataset, Class 2)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: SIGNALING PROTEIN |
 |
Structure Of Dock5/Elmo1/Rac1 Core (Rhog/Dock5/Elmo1/Rac1 Dataset, Class 3)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: SIGNALING PROTEIN |
 |
Structure Of Dock5/Elmo1/Rac1 Core (Rhog/Dock5/Elmo1/Rac1 Dataset, Class 4)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: SIGNALING PROTEIN |
 |
Structure Of Dock5/Elmo1/Rac1 Core (Rhog/Dock5/Elmo1/Rac1 Dataset, Class 5)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-29 Classification: SIGNALING PROTEIN |
 |
The Crystal Structure Of Cyanorhodopsin-Ii (Cyr-Ii) P7104R From Nodosilinea Nodulosa Pcc 7104
Organism: Nodosilinea nodulosa pcc 7104
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2023-10-25 Classification: MEMBRANE PROTEIN Ligands: RET, PG4, HEX, OCT, C14, R16, SO4, CL |
 |
Crystal Structure Of Hen Egg White Lysozyme Introduced With O-(2-Nitrobenzyl)-L-Tyrosine
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2022-09-21 Classification: BIOSYNTHETIC PROTEIN |
 |
Crystal Structure Of Photolysed Hen Egg White Lysozyme Introduced With O-(2-Nitrobenzyl)-L-Tyrosine
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2022-09-21 Classification: BIOSYNTHETIC PROTEIN |
 |
Crystal Structure Of O-(2-Nitrobenzyl)-L-Tyrosine-Trna Sythetase In Complex With O-(2-Nitrobenzyl)-L-Tyrosine
Organism: Methanocaldococcus jannaschii
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2022-09-21 Classification: TRANSCRIPTION Ligands: J2F |
 |
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-11-25 Classification: MEMBRANE PROTEIN Ligands: ZMA, NA, CLR, D12, MYS, HEX, 8K6, D10, OCT, UND, ER0, TRD |
 |
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-11-25 Classification: MEMBRANE PROTEIN Ligands: ZMA, NA, CLR, D12, MYS, HEX, 8K6, D10, OCT, UND, ER0, TRD |
 |
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-11-25 Classification: MEMBRANE PROTEIN Ligands: ZMA, NA, CLR, D12, MYS, HEX, 8K6, D10, OCT, UND, ER0, TRD |
 |
Crystal Structure Of Amp-Pnp Bound Mutant A3B3 Complex From Enterococcus Hirae V-Atpase
Organism: Enterococcus hirae (strain atcc 9790 / dsm 20160 / jcm 8729 / lmg 6399 / nbrc 3181 / ncimb 6459 / ncdo 1258)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-02-06 Classification: HYDROLASE Ligands: ANP, MG, GOL, MES |
 |
Organism: Enterococcus hirae (strain atcc 9790 / dsm 20160 / jcm 8729 / lmg 6399 / nbrc 3181 / ncimb 6459 / ncdo 1258)
Method: X-RAY DIFFRACTION Resolution:3.38 Å Release Date: 2019-02-06 Classification: HYDROLASE Ligands: GOL |
 |
Parallel Homodimer Structures Of Voltage-Gated Sodium Channel Beta4 For Cell-Cell Adhesion
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-07-05 Classification: CELL ADHESION Ligands: P33, PG4, GOL |
 |
Parallel Homodimer Structures Of The Extracellular Domains Of The Voltage-Gated Sodium Channel Beta4 Subunit Explain Its Role In Cell-Cell Adhesion
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2017-07-05 Classification: CELL ADHESION Ligands: GOL |

