Search Count: 230
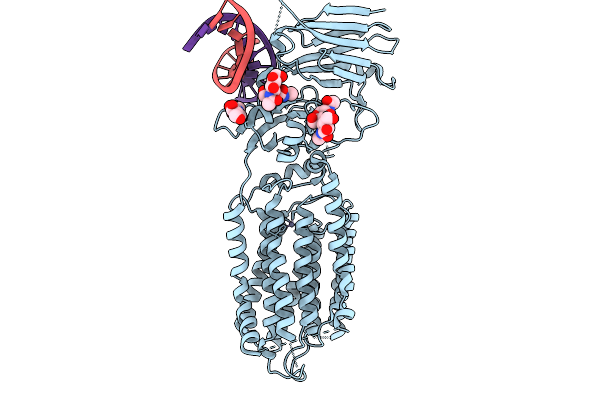 |
Organism: Caenorhabditis elegans, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.99 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN/RNA Ligands: NAG, ZN |
 |
Organism: Caenorhabditis elegans, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: MEMBRANE PROTEIN/RNA Ligands: NAG, ZN, Y01, LBN |
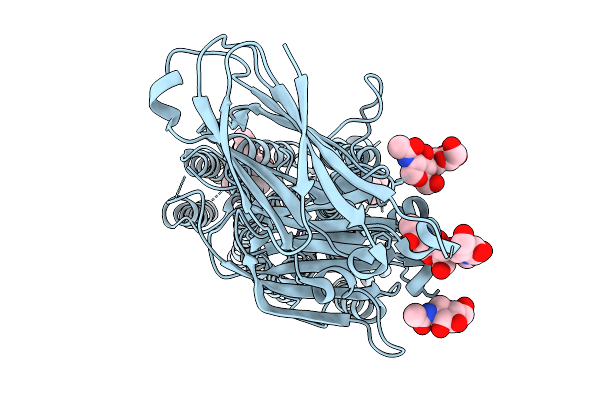 |
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2026-01-21 Classification: MEMBRANE PROTEIN Ligands: NAG, ZN, Y01, LBN |
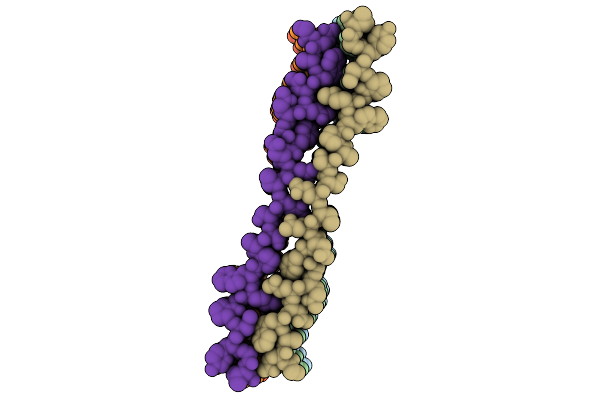 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: PROTEIN FIBRIL |
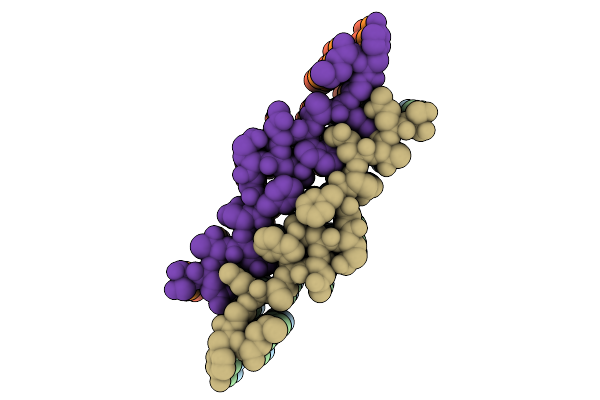 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: PROTEIN FIBRIL |
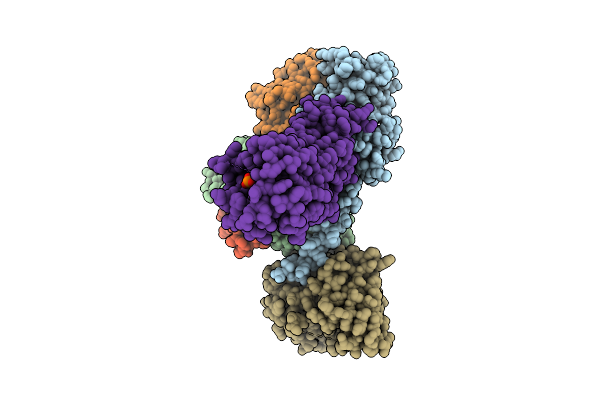 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.73 Å Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: A1L69 |
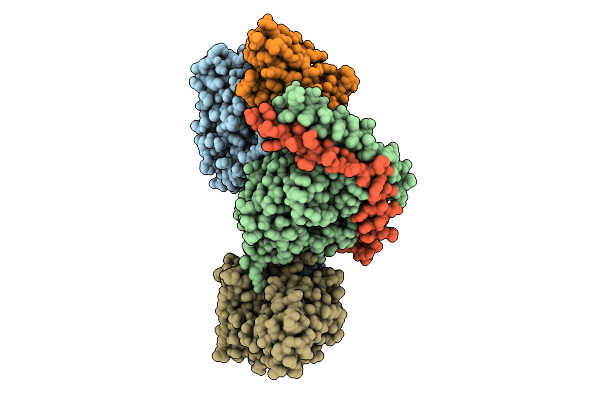 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.25 Å Release Date: 2025-11-26 Classification: MEMBRANE PROTEIN Ligands: S1P |
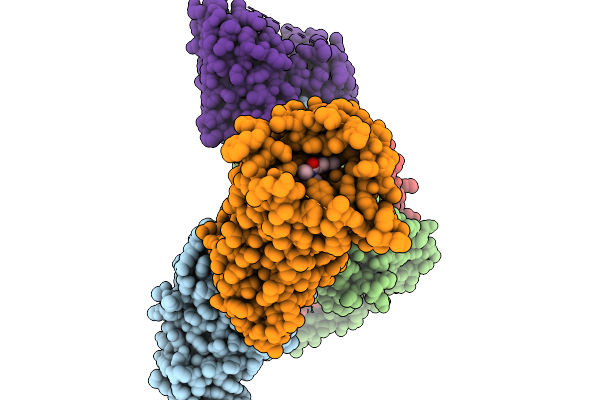 |
Cryo-Em Structure Of Human Kappa Opioid Receptor - G Protein Signaling Complex Bound With Nalfurafine.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.76 Å Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: IVB |
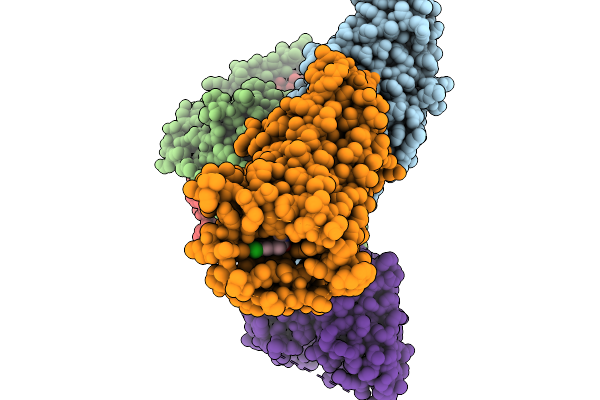 |
Cryo-Em Structure Of Human Kappa Opioid Receptor -G Protein Signaling Complex Bound With U-50488H
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: A1LWX |
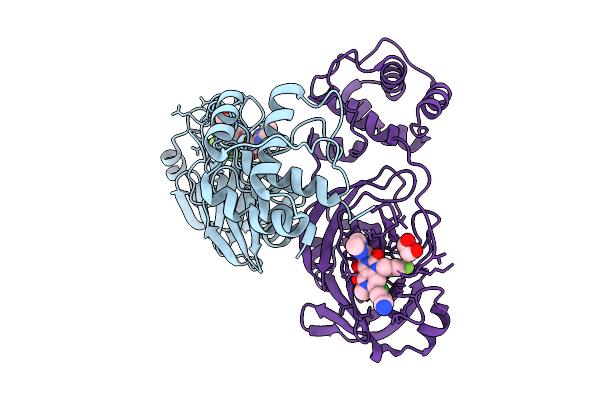 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-09-24 Classification: VIRAL PROTEIN Ligands: A1MA2, EDO |
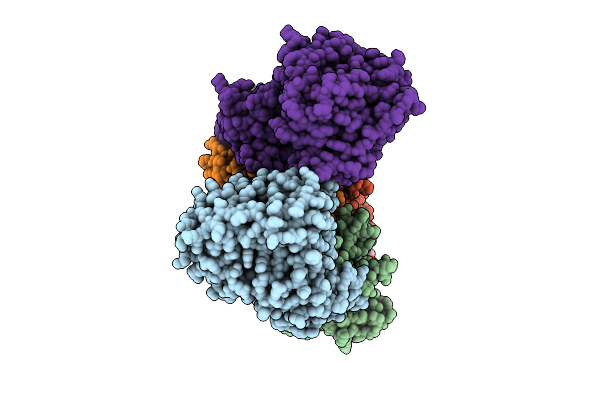 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, In The Absence Of Na+, Middle State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:3.75 Å Release Date: 2025-08-06 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, CA, LMT, FES, FAD |
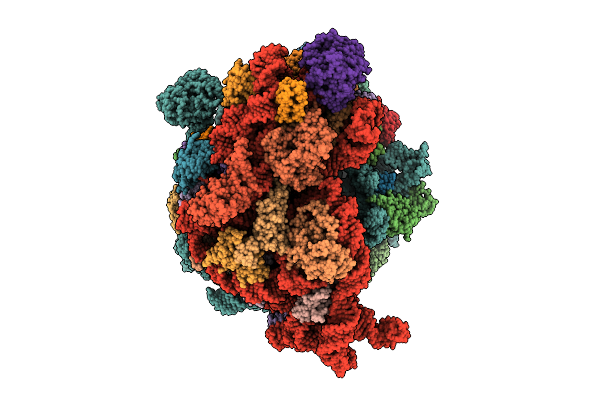 |
Cryo-Em Structure Of Escherichia Coli Hibernating Ribosome With Rnase I Mutant
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-07-30 Classification: RIBOSOME Ligands: MG, CA |
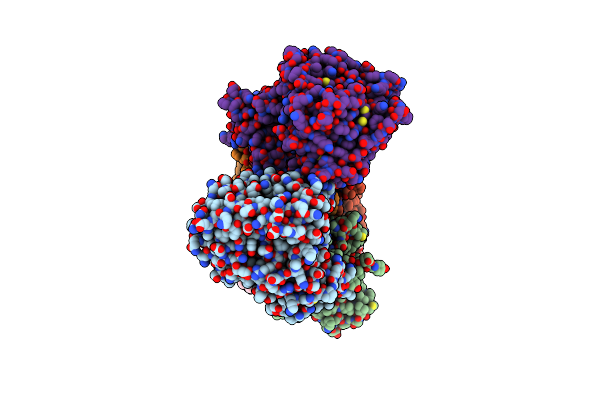 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrc-T225Y Mutant From Vibrio Cholerae
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:2.66 Å Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, LMT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrc-T225Y Mutant From Vibrio Cholerae Reduced By Nadh
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:2.59 Å Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, LMT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrb-T236Y Mutant From Vibrio Cholerae
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:3.80 Å Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: RBF, LMT, FMN, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrb-T236Y Mutant From Vibrio Cholerae Reduced By Nadh
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:3.31 Å Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: RBF, LMT, CA, FMN, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, With Bound Korormicin A
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, IQT, LMT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, In The Absence Of Na+, Upper State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:2.65 Å Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, LMT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, In The Absence Of Na+, Down State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:3.11 Å Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrb-G141A Mutant From Vibrio Cholerae Reduced By Nadh, With Bound Korormicin A, Stable State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:2.61 Å Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, IQT, CA, FES, FAD |

