Search Count: 141
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Resolution:3.19 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: 9UJ, A1LYC |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.36 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: A1LYD |
 |
Crystal Structure Of Measles Virus Fusion Inhibitor M1 Complexed With F Protein Hr1 (Hr1-42) (H32 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN Ligands: MG |
 |
Crystal Structure Of Measles Virus Fusion Inhibitor M1 Complexed With F Protein Hr1 (Hr1-42) (P21212 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:1.31 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN Ligands: MG |
 |
Crystal Structure Of Measles Virus Fusion Inhibitor M1 Complexed With F Protein Hr1 (Hr1-47) (P321 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN Ligands: CA |
 |
Crystal Structure Of Measles Virus Fusion Inhibitor M1Ek Complexed With F Protein Hr1 (Hr1-42) (P21 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN |
 |
Crystal Structure Of Measles Virus Fusion Inhibitor Mek28 Complexed With F Protein Hr1 (Hr1-40) (H3 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN Ligands: MPD |
 |
Crystal Structure Of Measles Virus Fusion Inhibitor Mek35Ge Complexed With F Protein Hr1 (Hr1-42) (P21212 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN Ligands: ZN, ACT |
 |
Crystal Structure Of Measles Virus Fusion Inhibitor Mek35Ge Complexed With F Protein Hr1 (Hr1-42) (P2 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN |
 |
Crystal Structure Of Measles Virus Fusion Inhibitor Mek35Gt Complexed With F Protein Hr1 (Hr1-42) (P21 Space Group)
Organism: Measles virus strain edmonston
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-01-01 Classification: ANTIVIRAL PROTEIN |
 |
Cryo-Em Structure Of The Potassium-Selective Channelrhodopsin Hckcr1 In Lipid Nanodisc
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Resolution:2.56 Å Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: RET, PSC, PLM |
 |
Cryo-Em Structure Of The Potassium-Selective Channelrhodopsin Hckcr2 In Lipid Nanodisc
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Resolution:2.53 Å Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: RET, PSC, PLM |
 |
Cryo-Em Structure Of The Potassium-Selective Channelrhodopsin Hckcr1 H225F Mutant In Lipid Nanodisc
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Resolution:2.66 Å Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: RET, PSC, PLM |
 |
Crystal Structure Of Bovine Heart Cytochrome C Oxidase, Apo Structure With Dmso
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-12-21 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PER, PGV, TGL, CUA, CDL, CHD, PEK, PSC, ZN, DMU |
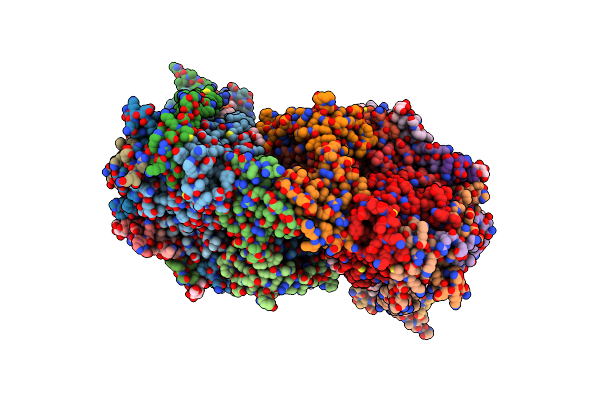 |
Crystal Structure Of Bovine Heart Cytochrome C Oxidase, The Structure Complexed With An Allosteric Inhibitor T113
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-12-21 Classification: OXIDOREDUCTASE Ligands: HEA, CU, MG, NA, PER, TGL, J6X, CUA, CHD, PSC, PEK, PGV, CDL, DMU, ZN |
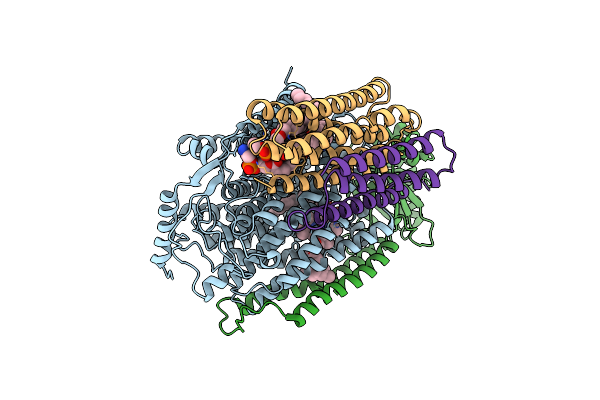 |
Cryo-Em Structure Of Cytochrome Bo3 From Escherichia Coli, Apo Structure With Dmso
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-12-21 Classification: OXIDOREDUCTASE Ligands: HEO, HEM, CU, PEE, UNX |
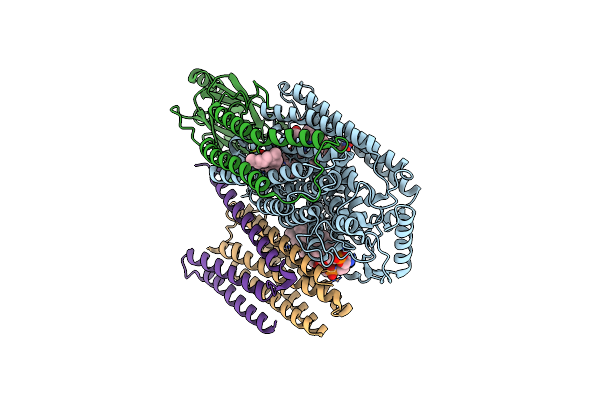 |
Cryo-Em Structure Of Cytochrome Bo3 From Escherichia Coli, The Structure Complexed With An Allosteric Inhibitor N4
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-12-21 Classification: OXIDOREDUCTASE Ligands: HEO, HEM, CU, PEE, JYR, UNX |
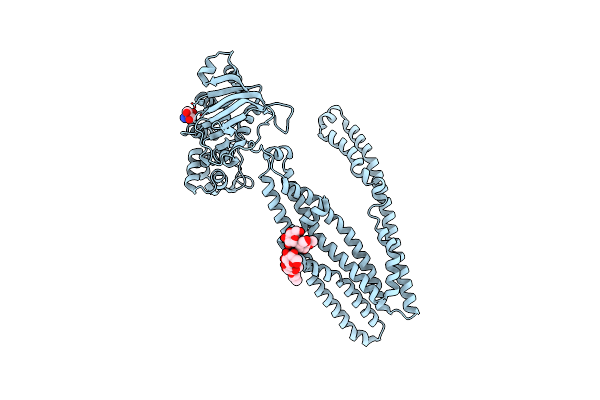 |
Crystal Structure Of Cmabcb1 W114Y/W161Y/W363Y/W364Y/M391W (4Wy/M391W) Mutant
Organism: Cyanidioschyzon merolae (strain 10d)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2022-09-07 Classification: TRANSPORT PROTEIN Ligands: DMU, TRS |
 |
Organism: Homo sapiens, Rattus norvegicus, Bos taurus, Unidentified
Method: ELECTRON MICROSCOPY Release Date: 2022-08-03 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens, Rattus norvegicus, Bos taurus, Unidentified
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2022-08-03 Classification: SIGNALING PROTEIN |

