Search Count: 36
 |
Cryo-Em Structure Of A 1033 Scaffold Base Dna Origami Nanostructure V4 And Tba
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: DNA |
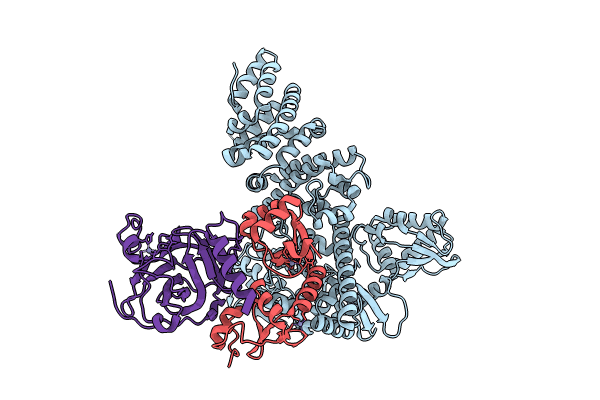 |
Organism: Homo sapiens, Saccharomyces cerevisiae (strain atcc 204508 / s288c), Human papillomavirus type 16
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: LIGASE Ligands: ZN |
 |
Organism: Homo sapiens, Saccharomyces cerevisiae s288c, Human papillomavirus type 16
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: LIGASE Ligands: ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: LIGASE Ligands: ZN |
 |
 |
Cryo-Em Structure Of Clock-Bmal1 Bound To A Nucleosomal E-Box At Position Shl-6.2 (Dna Conformation 1)
Organism: Homo sapiens, Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: GENE REGULATION |
 |
Cryo-Em Structure Of Clock-Bmal1 Bound To A Nucleosomal E-Box At Position Shl+5.8 (Composite Map)
Organism: Homo sapiens, Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: GENE REGULATION |
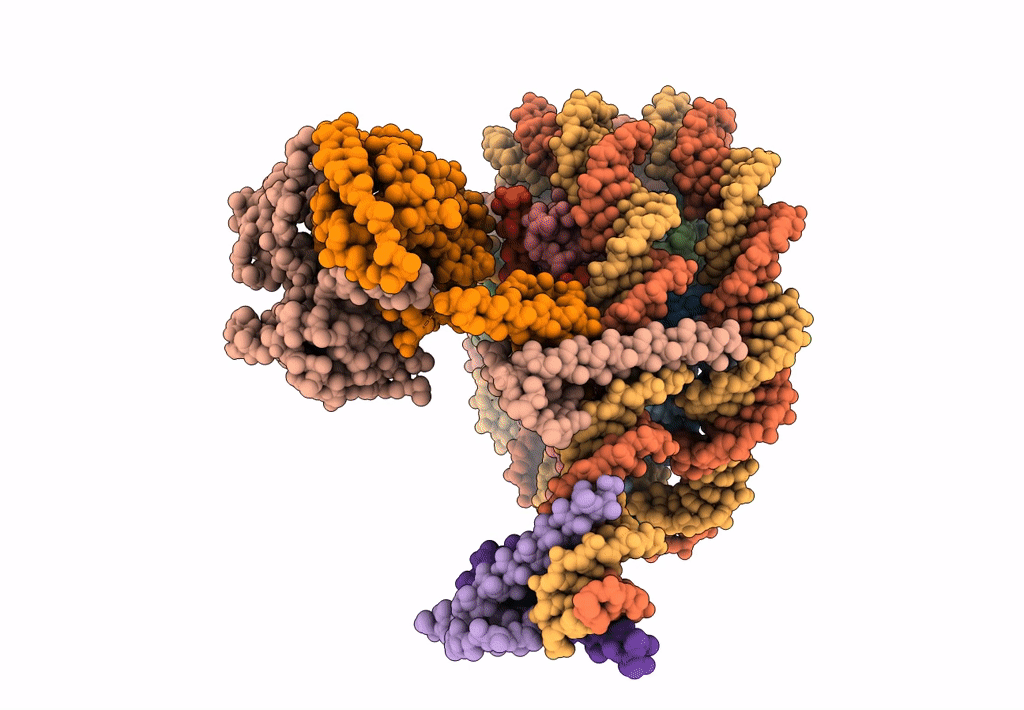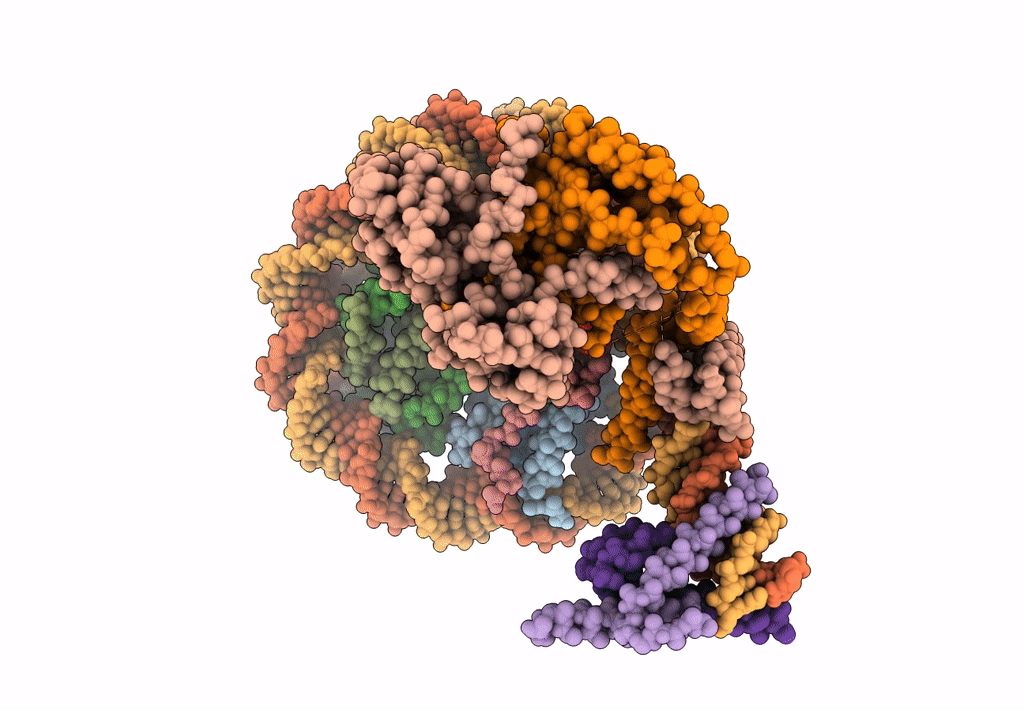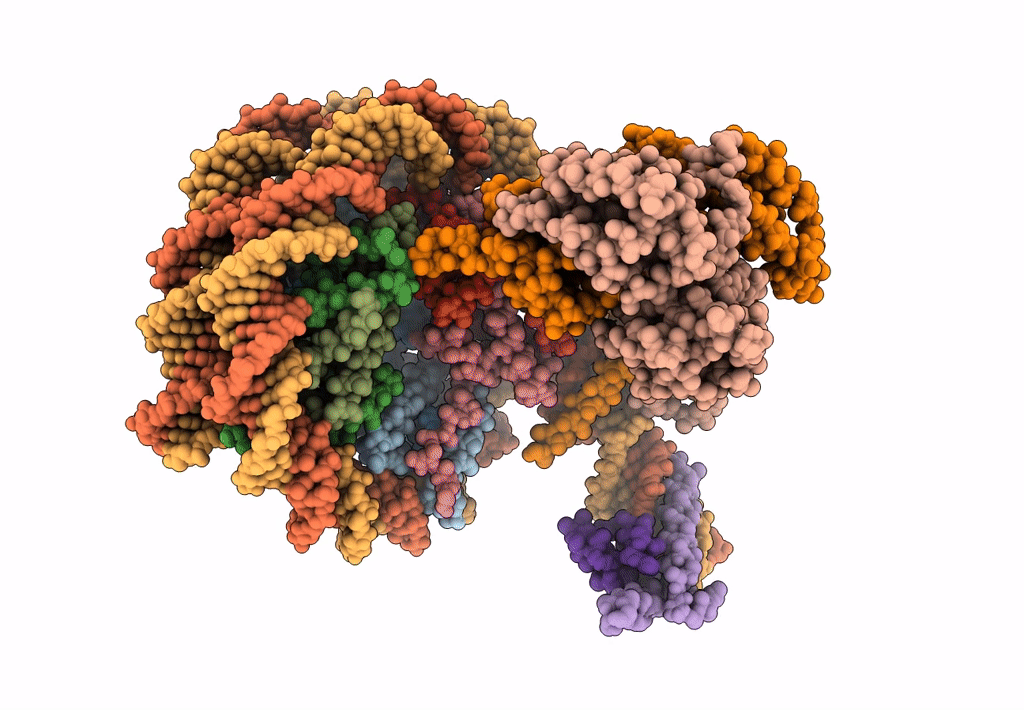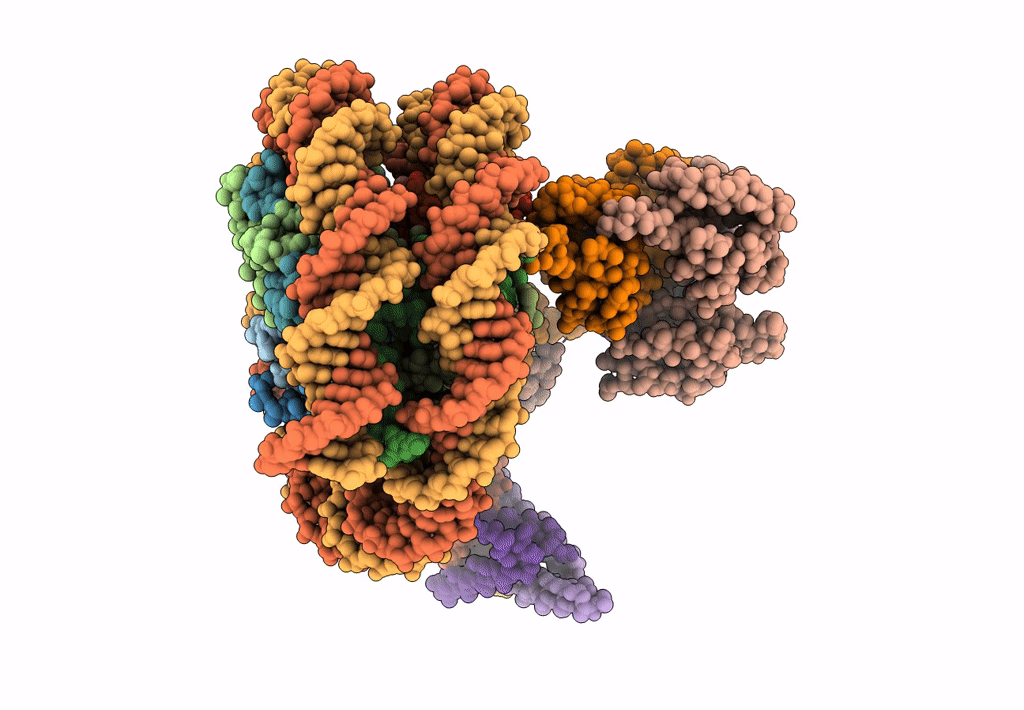 |
Cryo-Em Structure Of Clock-Bmal1 Bound To The Native Por Enhancer Nucleosome (Map 2, Additional 3D Classification And Flexible Refinement)
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: GENE REGULATION |
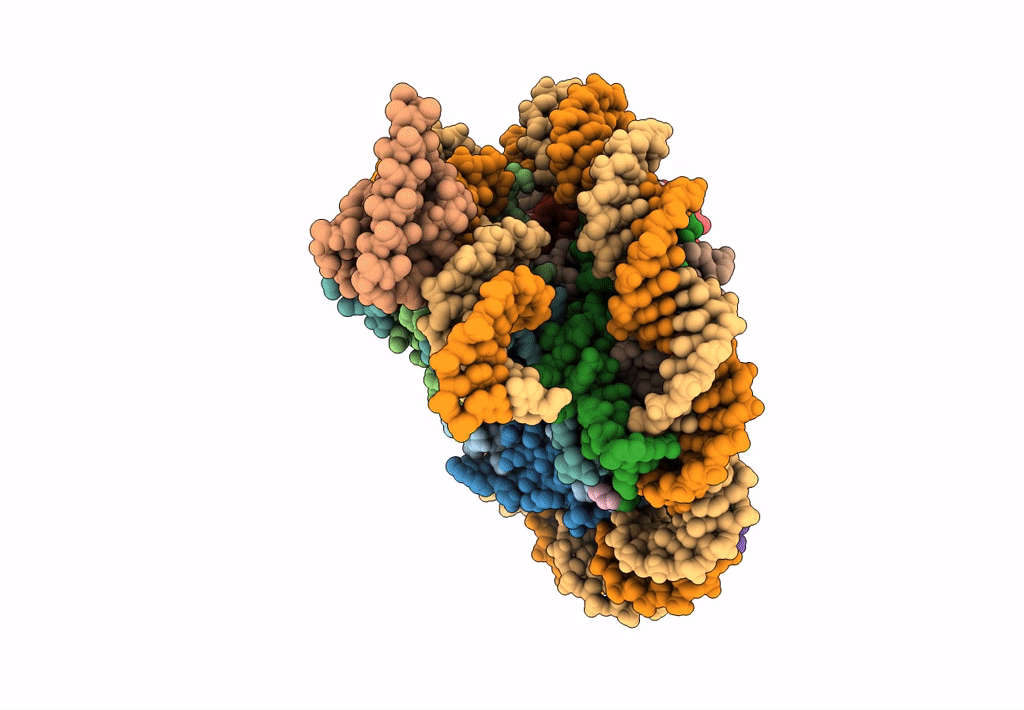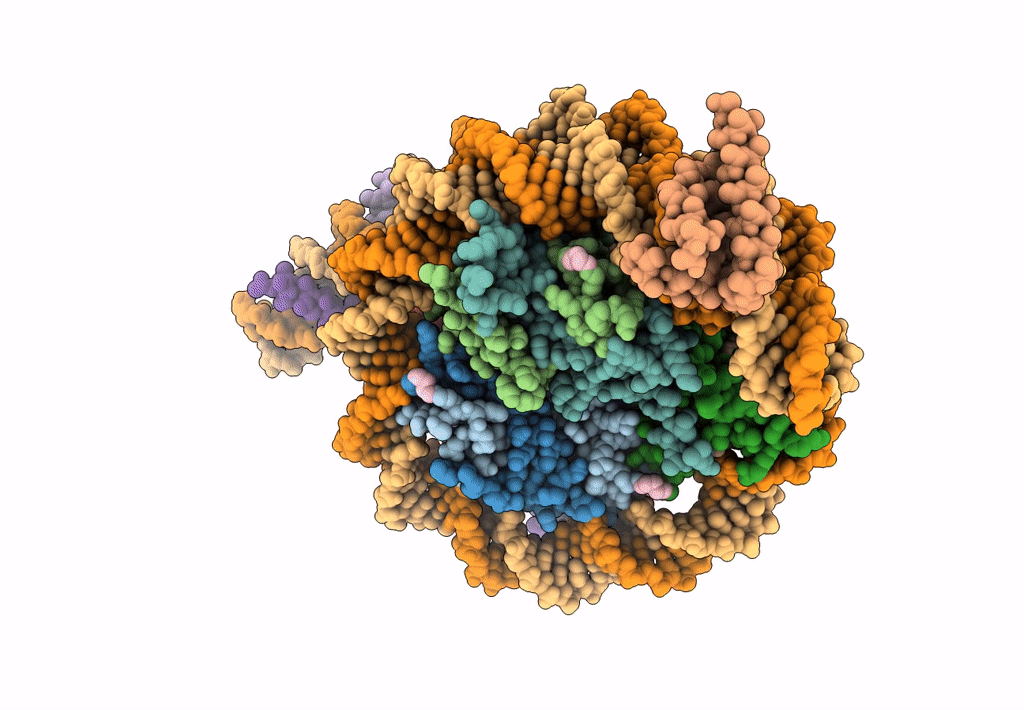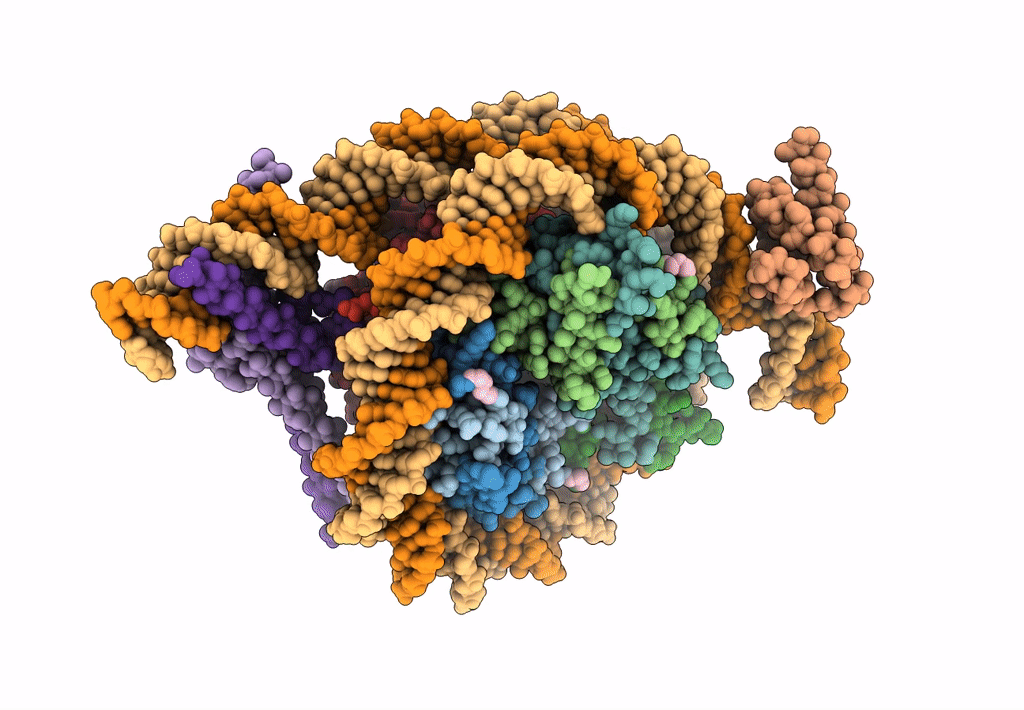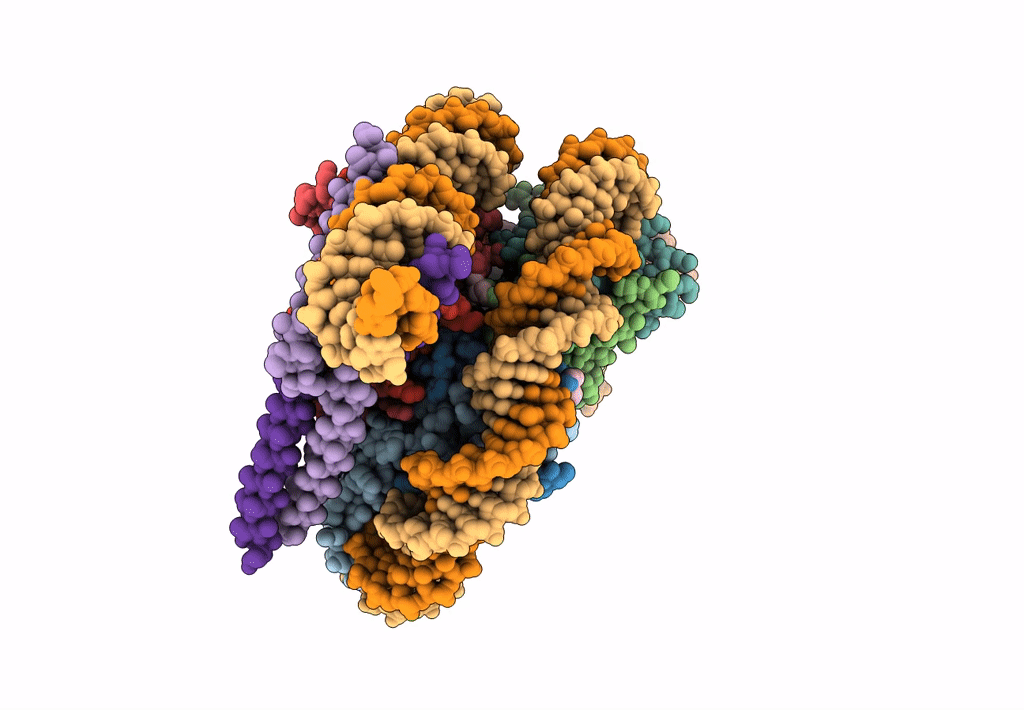 |
Organism: Homo sapiens, Aequorea victoria, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: TRANSCRIPTION Ligands: PTD |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: TRANSCRIPTION Ligands: PTD |
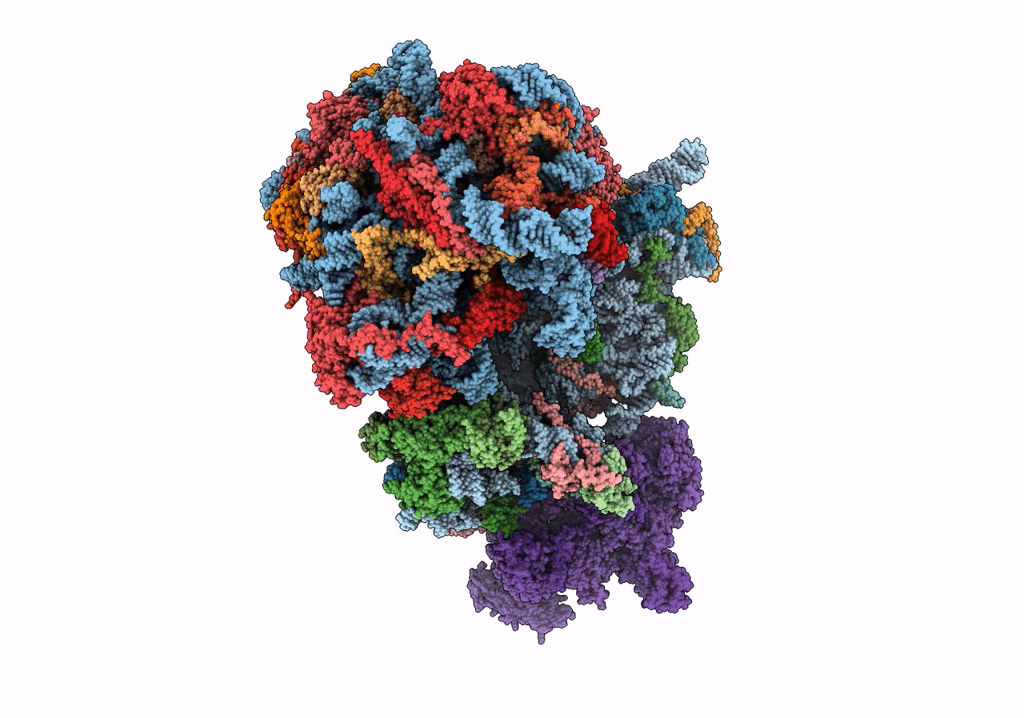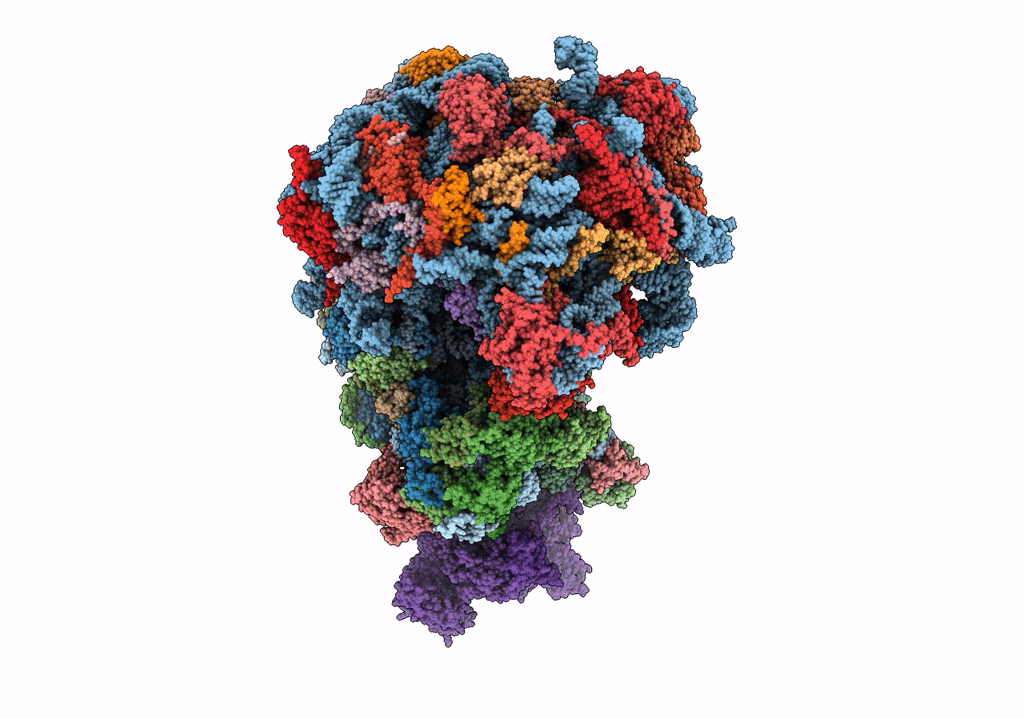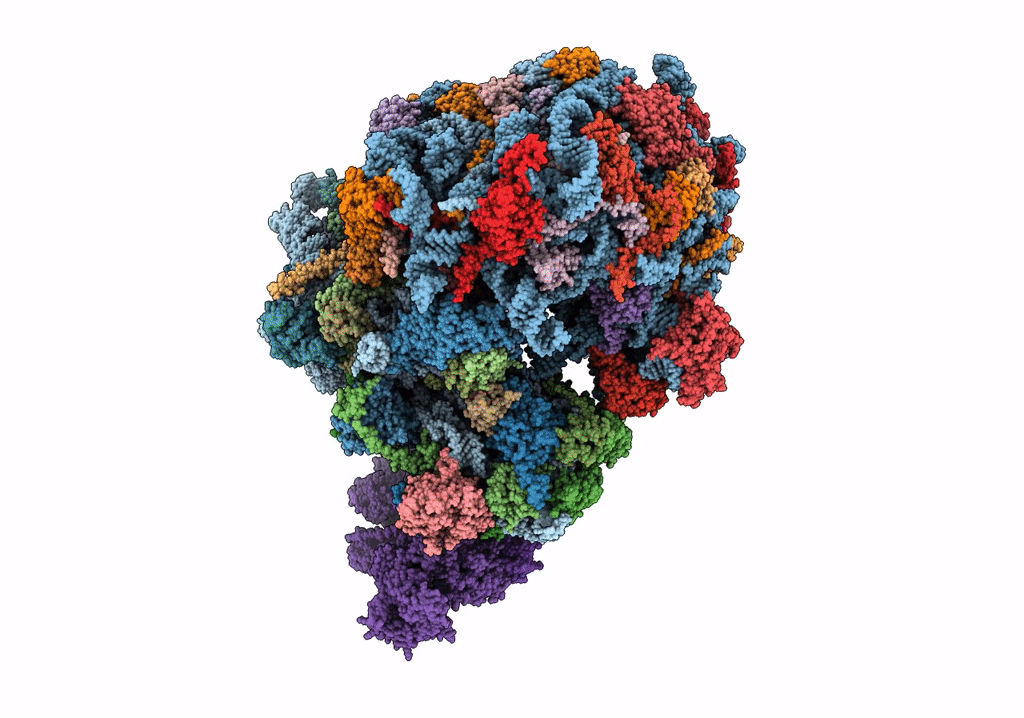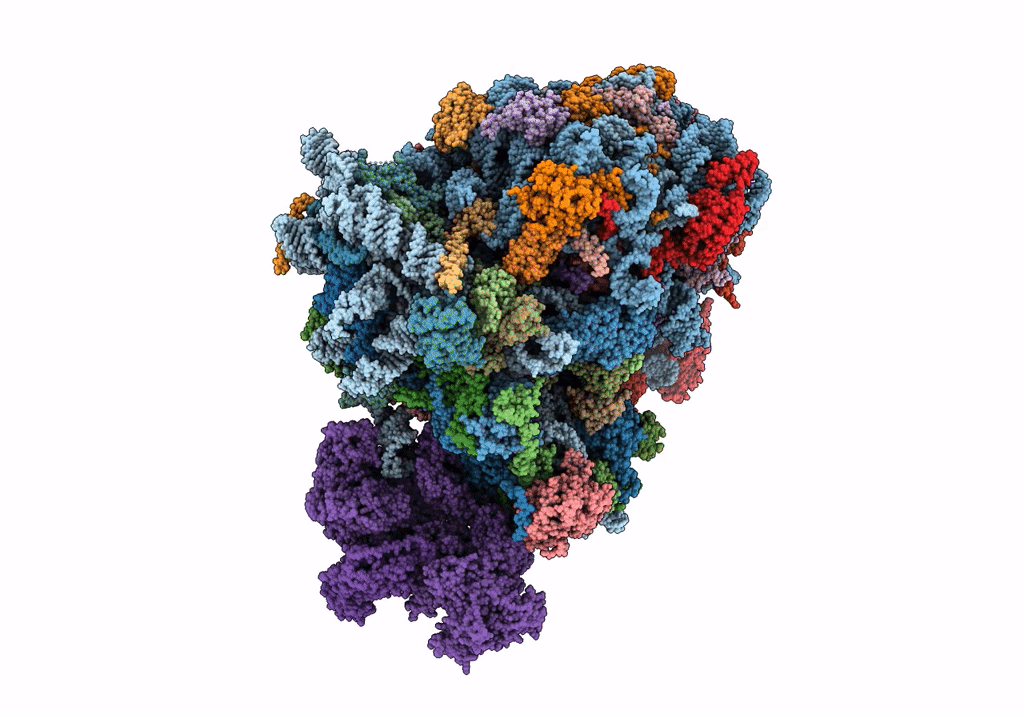 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-02-22 Classification: RIBOSOME Ligands: MG, ZN |
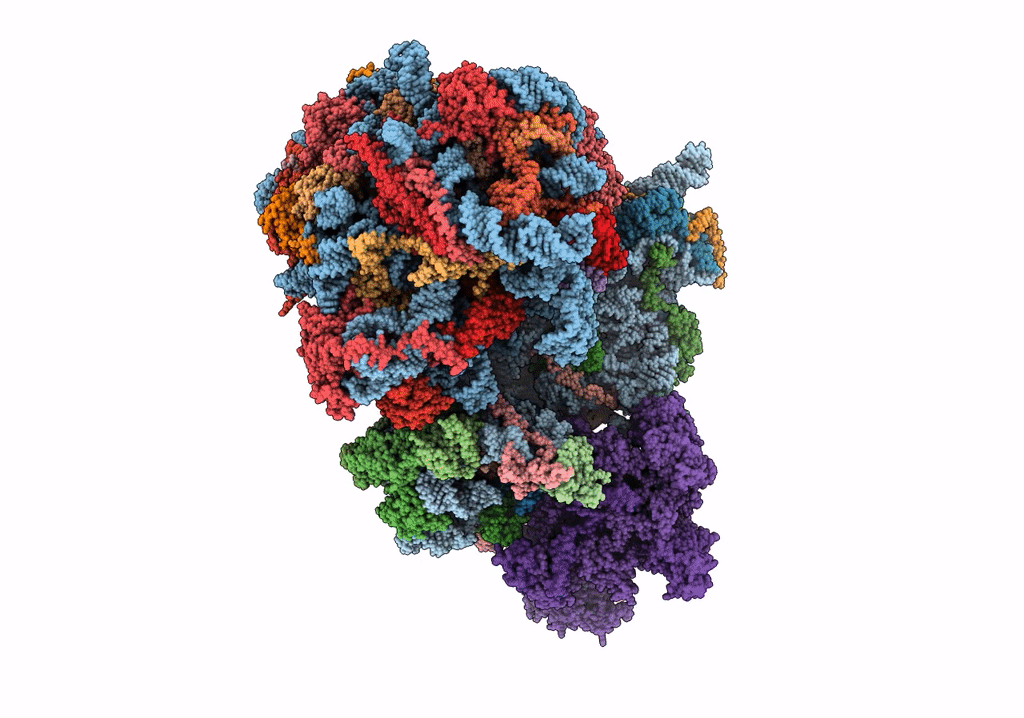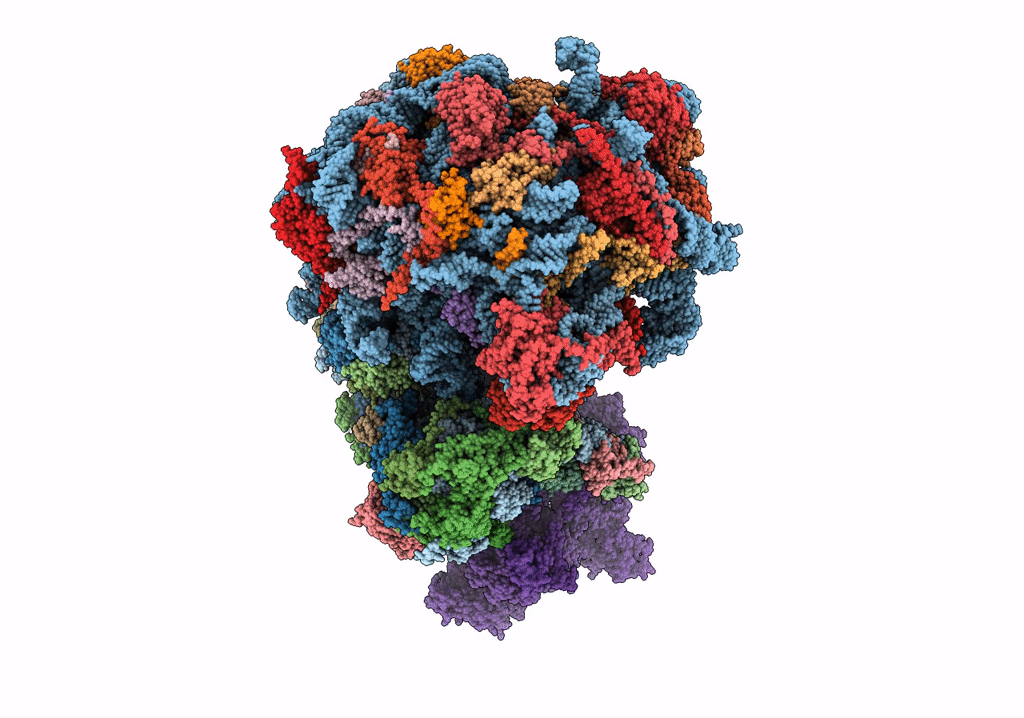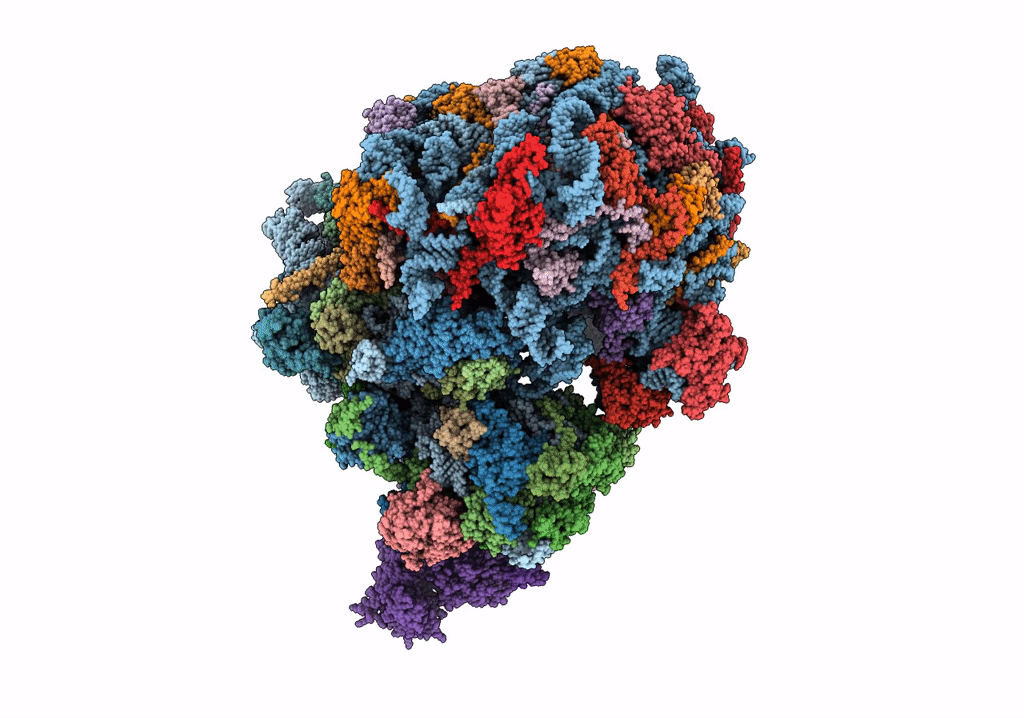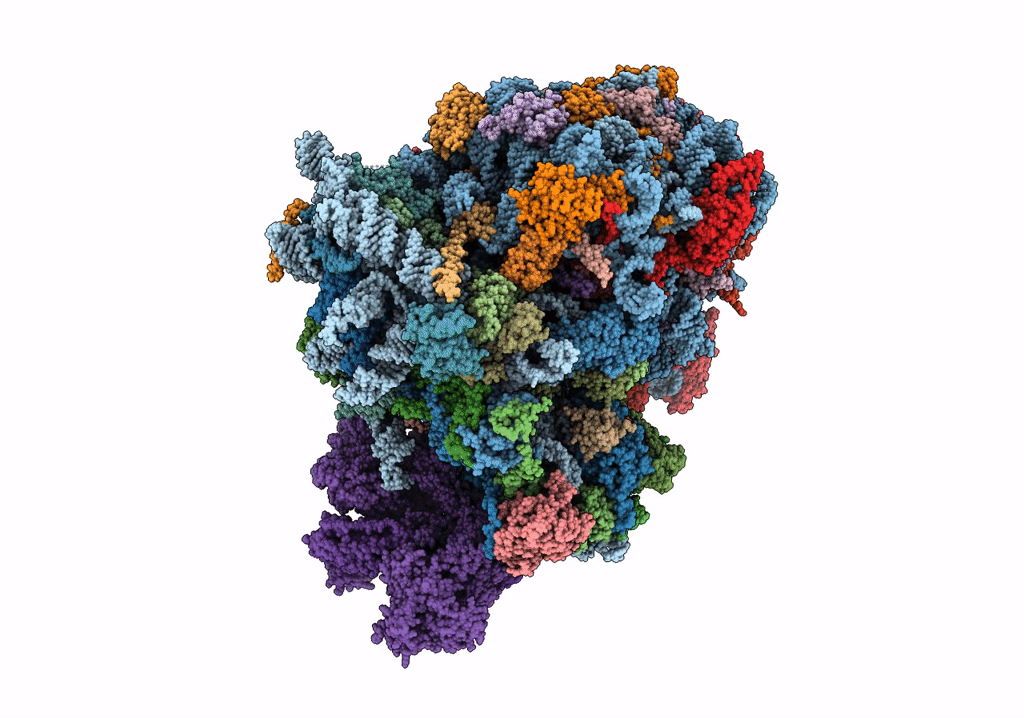 |
Structure Of The Rqt-Bound 80S Ribosome From S. Cerevisiae (C2) - Composite Map
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-02-22 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-02-22 Classification: RIBOSOME Ligands: MG |
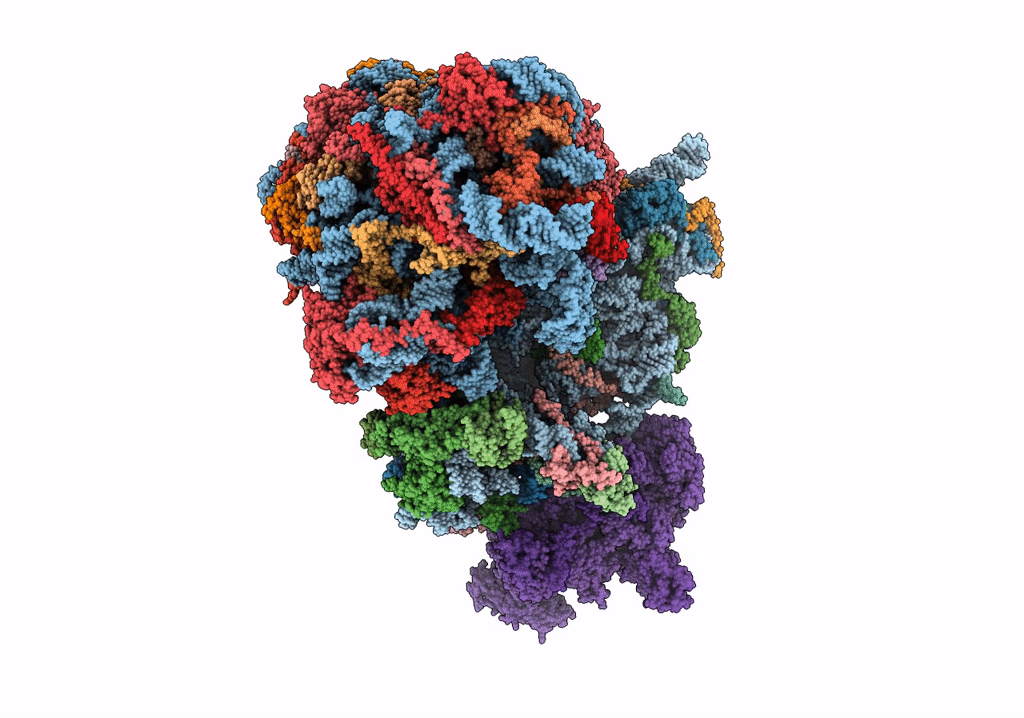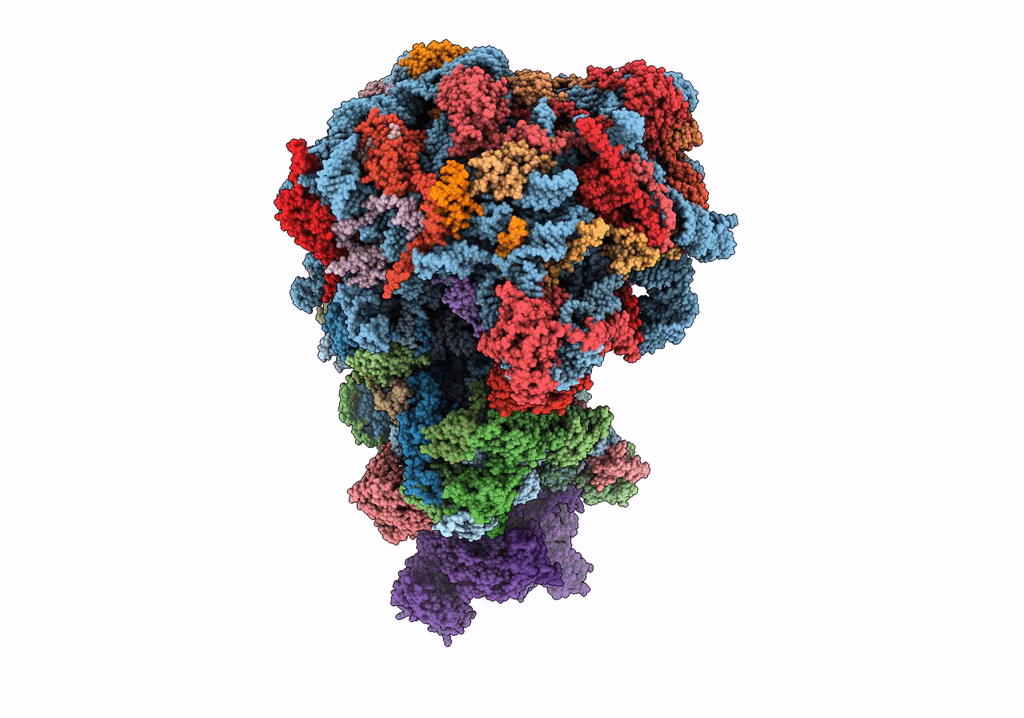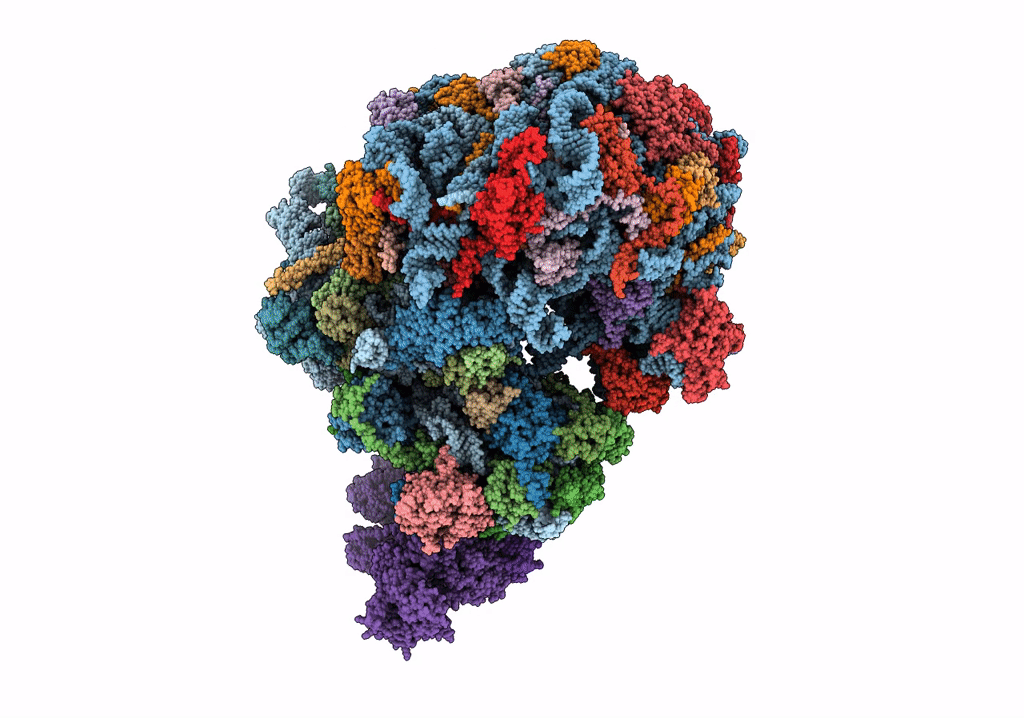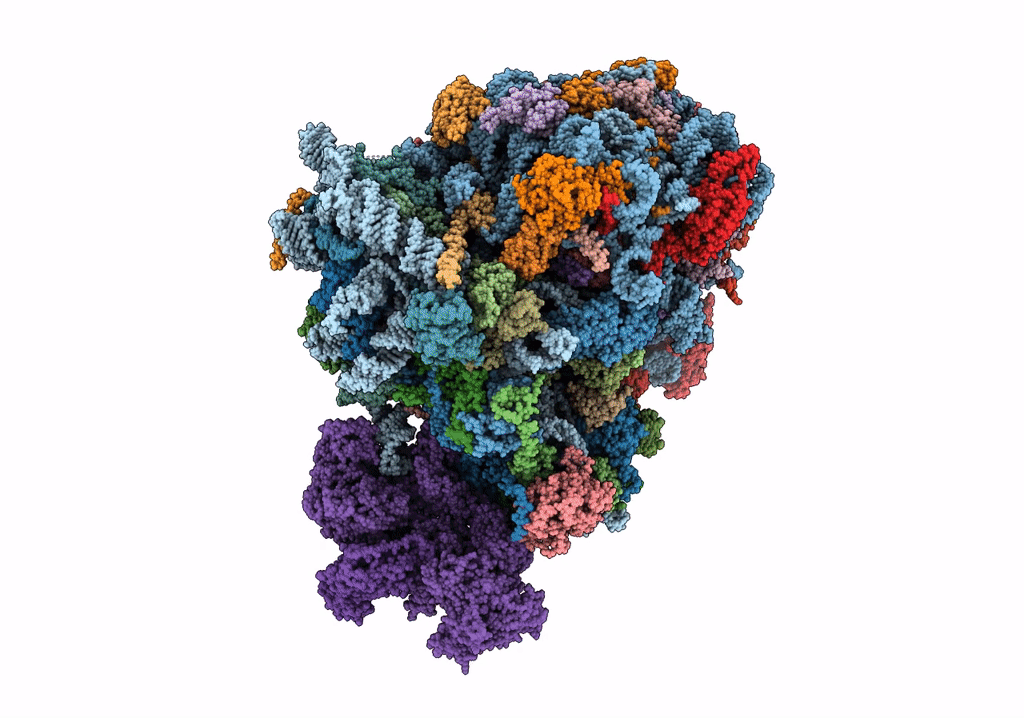 |
Structure Of Rqt (C1) Bound To The Stalled Ribosome In A Disome Unit From S. Cerevisiae
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-02-22 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-02-22 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-02-10 Classification: DNA BINDING PROTEIN |
 |
Rix1-Rea1 Pre-60S Particle - Rix1-Subcomplex, Body 3 (Rigid Body Refinement)
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2020-07-29 Classification: RIBOSOME |
 |
Rix1-Rea1 Pre-60S Particle - Rea1, Body 3 (Rigid Body Refinement, Composite Structure Of Rea1 Ring And Tail)
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2020-07-29 Classification: RIBOSOME |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2020-07-29 Classification: RIBOSOME Ligands: ZN, GTP, MG |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2020-07-29 Classification: RIBOSOME Ligands: ZN, GTP, MG |

