Search Count: 55
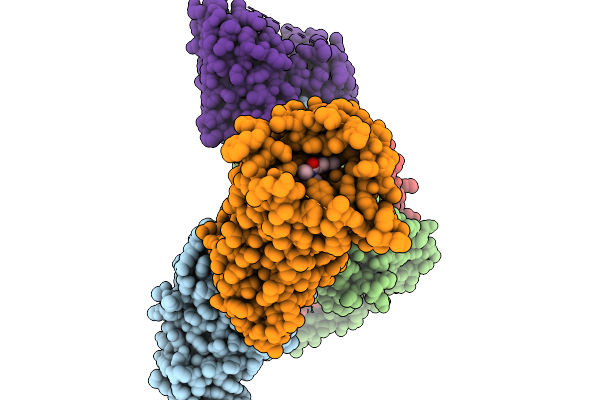 |
Cryo-Em Structure Of Human Kappa Opioid Receptor - G Protein Signaling Complex Bound With Nalfurafine.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: IVB |
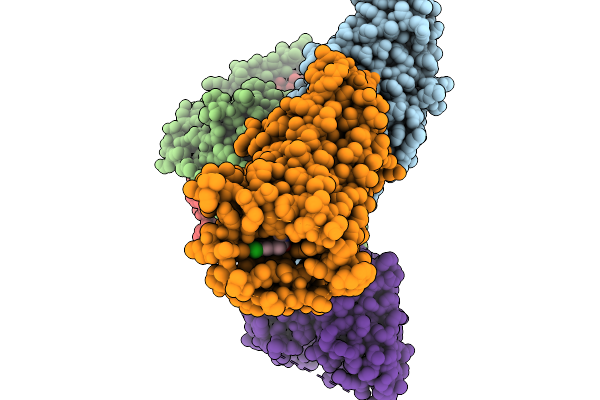 |
Cryo-Em Structure Of Human Kappa Opioid Receptor -G Protein Signaling Complex Bound With U-50488H
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN Ligands: A1LWX |
 |
Crystal Structure Of The Light-Driven Proton Pump Heimdallarchaeial Rhodopsin Heimdallr1
Organism: Candidatus heimdallarchaeota archaeon
Method: X-RAY DIFFRACTION Release Date: 2025-04-02 Classification: PROTON TRANSPORT Ligands: RET, OLC, NO3 |
 |
Organism: Guillardia theta ccmp2712
Method: ELECTRON MICROSCOPY Resolution:2.86 Å Release Date: 2024-09-04 Classification: MEMBRANE PROTEIN Ligands: RET |
 |
Organism: Guillardia theta ccmp2712
Method: ELECTRON MICROSCOPY Resolution:2.73 Å Release Date: 2024-09-04 Classification: MEMBRANE PROTEIN Ligands: RET, PC1 |
 |
Organism: Guillardia theta
Method: ELECTRON MICROSCOPY Resolution:2.71 Å Release Date: 2024-09-04 Classification: MEMBRANE PROTEIN Ligands: RET, PC1 |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Resolution:2.70 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: A1L0C |
 |
Organism: Canis lupus familiaris
Method: ELECTRON MICROSCOPY Resolution:2.20 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: A1L0D |
 |
Organism: Canis lupus familiaris
Method: ELECTRON MICROSCOPY Resolution:2.00 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: A1L0C |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Resolution:2.50 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: A1L0D, THR |
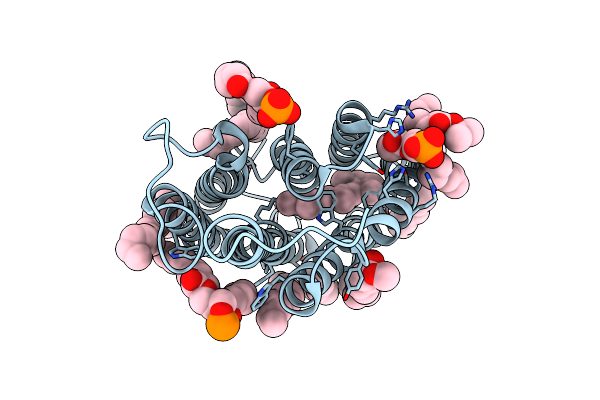 |
Cryo-Em Structure Of The Potassium-Selective Channelrhodopsin Hckcr1 In Lipid Nanodisc
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Resolution:2.56 Å Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: RET, PSC, PLM |
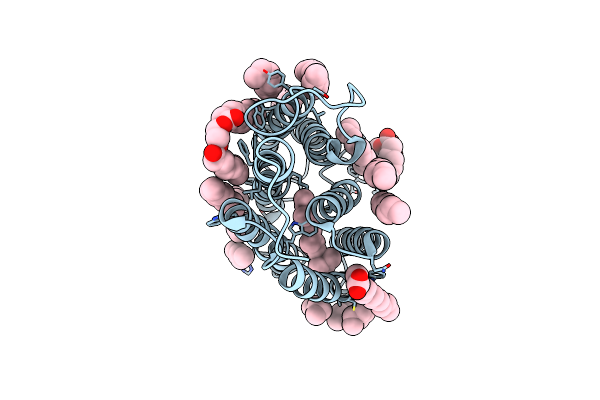 |
Cryo-Em Structure Of The Potassium-Selective Channelrhodopsin Hckcr2 In Lipid Nanodisc
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Resolution:2.53 Å Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: RET, PSC, PLM |
 |
Cryo-Em Structure Of The Potassium-Selective Channelrhodopsin Hckcr1 H225F Mutant In Lipid Nanodisc
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Resolution:2.66 Å Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: RET, PSC, PLM |
 |
Organism: Lama glama, Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.70 Å Release Date: 2023-08-30 Classification: MEMBRANE PROTEIN Ligands: RET |
 |
Organism: Lama glama, Bos taurus
Method: X-RAY DIFFRACTION Resolution:3.71 Å Release Date: 2023-08-30 Classification: MEMBRANE PROTEIN |
 |
Organism: Lama glama, Bos taurus
Method: X-RAY DIFFRACTION Resolution:4.25 Å Release Date: 2023-08-30 Classification: MEMBRANE PROTEIN |
 |
Organism: Uncultured bdellovibrionales bacterium
Method: ELECTRON MICROSCOPY Release Date: 2023-03-29 Classification: PROTON TRANSPORT Ligands: RET, K3I |
 |
Organism: Bdellovibrio
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2023-03-15 Classification: MEMBRANE PROTEIN Ligands: RET, OLC |
 |
Cryo-Em Structure Of Bestrhodopsin (Rhodopsin-Rhodopsin-Bestrophin) Complex
Organism: Phaeocystis
Method: ELECTRON MICROSCOPY Release Date: 2022-07-06 Classification: MEMBRANE PROTEIN Ligands: RET |
 |
Organism: Sapovirus hu/gi.6/nichinan/fp05284/2005/jpn
Method: ELECTRON MICROSCOPY Release Date: 2021-12-15 Classification: VIRUS |

