Search Count: 37
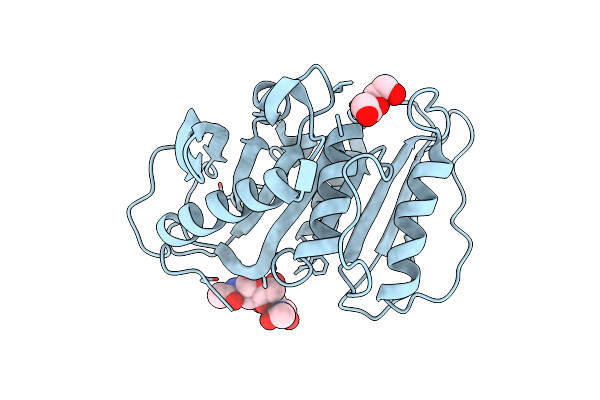 |
Structure-Based Characterization And Improvement Of An Enzymatic Activity Of Acremonium Alcalophilum Feruloyl Esterase
Organism: Sodiomyces alcalophilus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-04-17 Classification: HYDROLASE Ligands: PEG, MG |
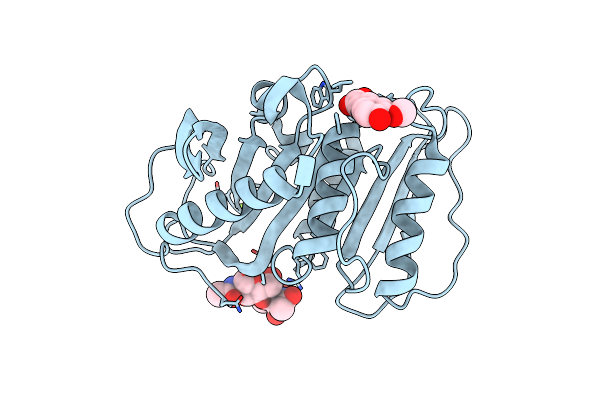 |
Structure-Based Characterization And Improvement Of An Enzymatic Activity Of Acremonium Alcalophilum Feruloyl Esterase
Organism: Sodiomyces alcalophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-04-17 Classification: HYDROLASE Ligands: FER, MG |
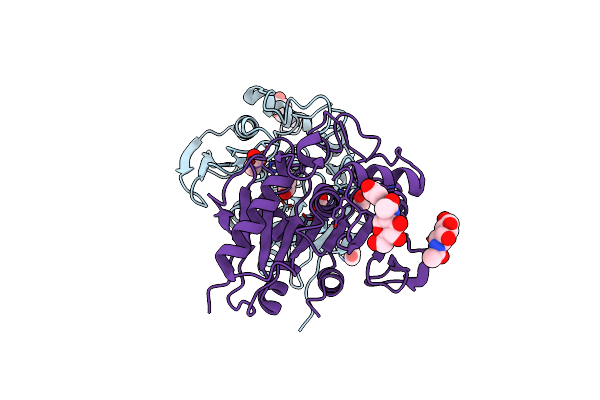 |
Structure Insight Into Substrate Recognition And Catalysis By Feruloyl Esterase From Aspergillus Sydowii
Organism: Aspergillus sydowii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-10-25 Classification: HYDROLASE Ligands: NAG, ACT |
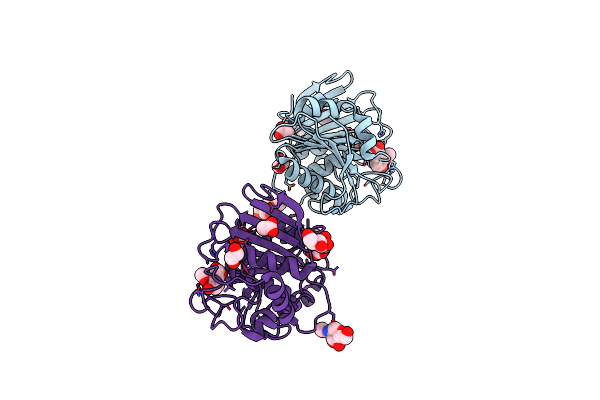 |
Structure Insight Into Substrate Recognition And Catalysis By Feruloyl Esterase From Aspergillus Sydowii
Organism: Aspergillus sydowii
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2023-10-25 Classification: HYDROLASE Ligands: NAG, FER, ACT, EDO |
 |
Structure Insight Into Substrate Recognition And Catalysis By Feruloyl Esterase From Aspergillus Sydowii
Organism: Aspergillus sydowii
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-10-25 Classification: HYDROLASE Ligands: SXX, NAG, EDO, ACT |
 |
Organism: Ceriporiopsis subvermispora (strain b)
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2022-05-04 Classification: OXIDOREDUCTASE Ligands: NAG, CU, TRS, EDO, XYS, CA |
 |
 |
Crystal Structure Of An Apo Form Of The Glutathione S-Transferase, Csgst83044, Of Ceriporiopsis Subvermispora
Organism: Ceriporiopsis subvermispora (strain b)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2019-05-15 Classification: TRANSFERASE Ligands: EDO, CA, SO4 |
 |
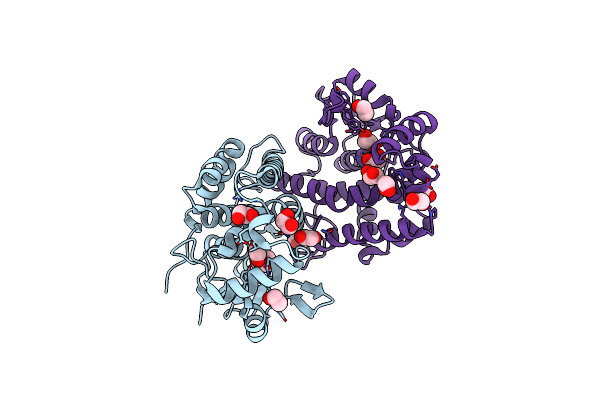
Crystal Structure Of The Glutathione S-Transferase, Csgst83044, Of Ceriporiopsis Subvermispora In Complex With Glutathione
Organism: Ceriporiopsis subvermispora (strain b)
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2019-05-15 Classification: TRANSFERASE Ligands: GSH |
 |
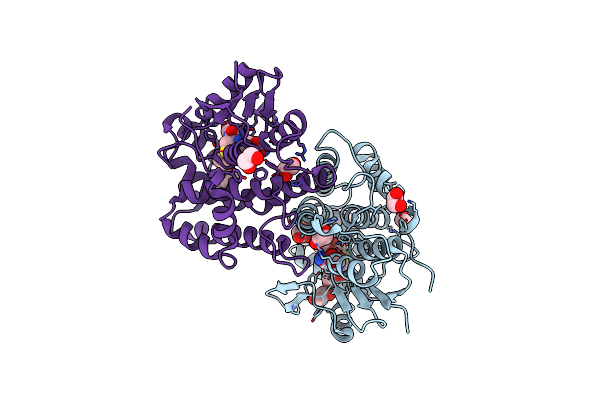
Crystal Structure Of An Apo Form Of The Glutathione S-Transferase, Csgst63524, Of Ceriporiopsis Subvermispora
Organism: Ceriporiopsis subvermispora (strain b)
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2019-02-27 Classification: TRANSFERASE Ligands: EDO |
 |
Crystal Structure Of The Glutathione S-Transferase, Csgst63524, Of Ceriporiopsis Subvermispora In Complex With Glutathione
Organism: Ceriporiopsis subvermispora (strain b)
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-02-27 Classification: TRANSFERASE Ligands: GSH, EDO |
 |
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 2017-12-13 Classification: RNA BINDING PROTEIN |
 |
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 2017-12-13 Classification: RNA BINDING PROTEIN/RNA |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2014-10-15 Classification: PEPTIDE BINDING PROTEIN |
 |
Refined Structure Of Rna Aptamer In Complex With The Partial Binding Peptide Of Prion Protein
Organism: Bos taurus
Method: SOLUTION NMR Release Date: 2014-05-21 Classification: Membrane Protein/RNA |
 |
Rna Aptamer Against Prion Protein In Complex With The Partial Binding Peptide
Organism: Bos taurus
Method: SOLUTION NMR Release Date: 2013-02-13 Classification: Membrane Protein/RNA |
 |
1H, 13C, And 15N Chemical Shift Assignments For Musashi1 Rbd1:R(Guagu) Complex
Organism: Mus musculus
Method: SOLUTION NMR Release Date: 2011-12-28 Classification: RNA BINDING PROTEIN/RNA |
 |
 |
Structure, Interaction, And Real-Time Monitoring Of The Enzymatic Reaction Of Wild Type Apobec3G
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2009-02-03 Classification: HYDROLASE Ligands: ZN |
 |
Solution Structure Of Fully Modified 4'-Thiodna With The Sequence Of D(Cgcgaattcgcg)
|

