Search Count: 23
 |
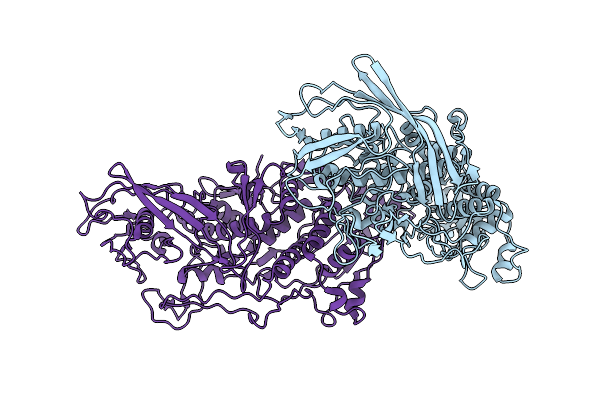
Cryo-Em Reconstruction Of The Full-Length E. Coli Transmembrane Formate Transporter Foca
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN Ligands: HOH |
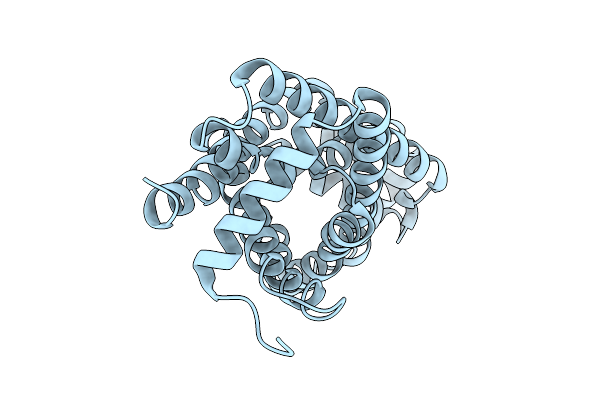 |
Cryo-Em Reconstruction Of The Full-Length H209N Mutant E. Coli Transmembrane Formate Transporter Foca
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN |
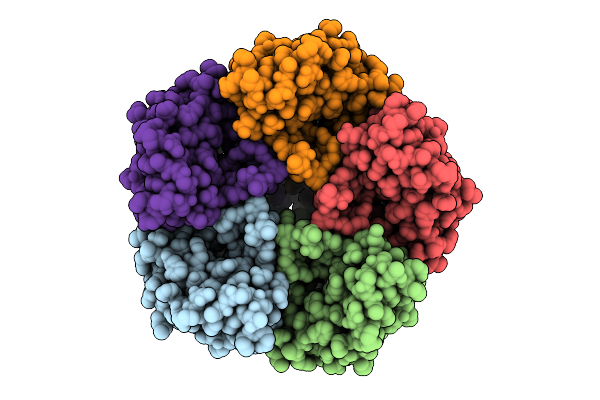 |
Asymmetric Cryo-Em Reconstruction Of The Full-Length E. Coli Transmembrane Formate Transporter Foca
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN |
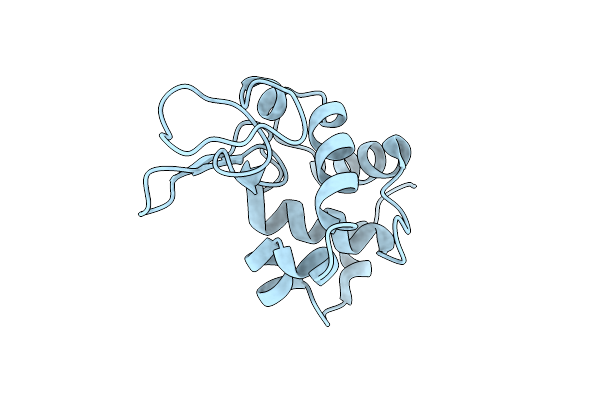 |
Hen Egg-White Lysozyme Structure Determined By 3Ded/Microed On A 200 Kev Microscope
Organism: Gallus gallus
Method: ELECTRON CRYSTALLOGRAPHY Resolution:2.80 Å Release Date: 2025-03-19 Classification: HYDROLASE |
 |
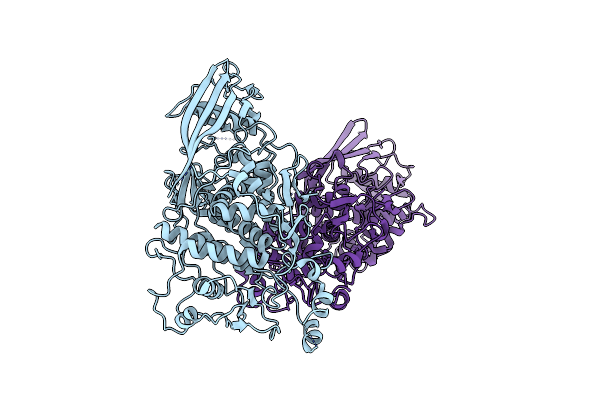
Myo-Inositol-1-Phosphate Synthase From Thermochaetoides Thermophila In Complex With Nad
Organism: Thermochaetoides thermophila dsm 1495
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: ISOMERASE Ligands: NAD |
 |
Organism: Saccharomyces cerevisiae virus l-a
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: VIRUS |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: VIRUS |
 |
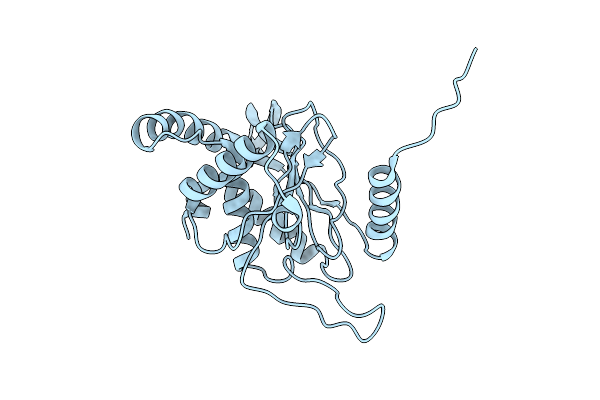
Symmetry Expanded D7 Local Refined Map Of Mitochondrial Heat-Shock Protein 60-Like Protein From Chaetomium Thermophilum
Organism: Thermochaetoides thermophila
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: CHAPERONE |
 |
Cryo-Em Structure Of Photosystem Ii C2S2 Supercomplex From Norway Spruce (Picea Abies) At 2.8 Angstrom Resolution
|
 |
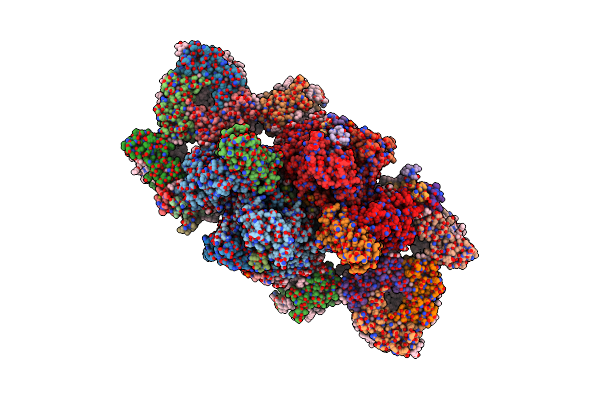
Cryo-Em Reconstruction Of The Native 24-Mer E2O Core Of The 2-Oxoglutarate Dehydrogenase Complex Of C. Thermophilum At 3.35 A Resolution
Organism: Thermochaetoides thermophila dsm 1495
Method: ELECTRON MICROSCOPY Release Date: 2023-05-31 Classification: TRANSFERASE |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-11-02 Classification: SIGNALING PROTEIN Ligands: ZN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-11-02 Classification: SIGNALING PROTEIN |
 |
The Structure Of Photosystem I Tetramer From Chroococcidiopsis Ts-821, A Thermophilic, Unicellular, Non-Heterocyst-Forming Cyanobacterium
Organism: Chroococcidiopsis sp. ts-821
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: PHOTOSYNTHESIS Ligands: LHG, CLA, PQN, BCR, SF4 |
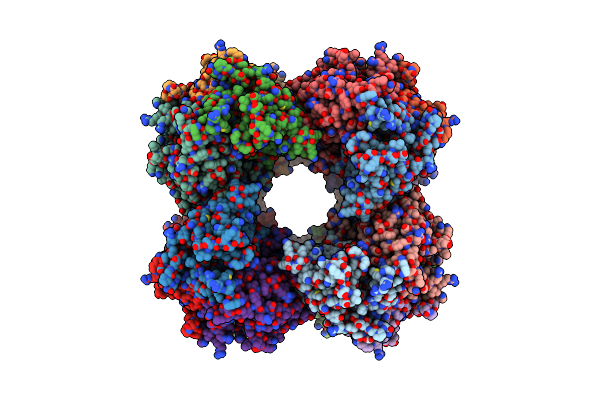 |
Protein Community Member Oxoglutarate Dehydrogenase Complex E2 Core From C. Thermophilum
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: ELECTRON MICROSCOPY Release Date: 2022-02-02 Classification: TRANSFERASE |
 |
Protein Community Member Pyruvate Dehydrogenase Complex E2 Core From C. Thermophilum
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: ELECTRON MICROSCOPY Release Date: 2022-02-02 Classification: TRANSFERASE |
 |
Organism: Chaetomium thermophilum var. thermophilum dsm 1495
Method: ELECTRON MICROSCOPY Release Date: 2022-02-02 Classification: TRANSFERASE |
 |
Metabolon-Embedded Pyruvate Dehydrogenase Complex E2 Core At Near-Atomic Resolution
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: ELECTRON MICROSCOPY Release Date: 2021-12-01 Classification: TRANSFERASE |
 |
Organism: Chaetomium thermophilum var. thermophilum dsm 1495
Method: ELECTRON MICROSCOPY Release Date: 2021-02-10 Classification: TRANSFERASE |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2020-08-26 Classification: RIBOSOME Ligands: ZN |
 |
Cryo-Em Structure Of K63 Ubiquitinated Yeast Translocating Ribosome Under Oxidative Stress
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2020-08-26 Classification: RIBOSOME Ligands: ZN |

