Search Count: 25
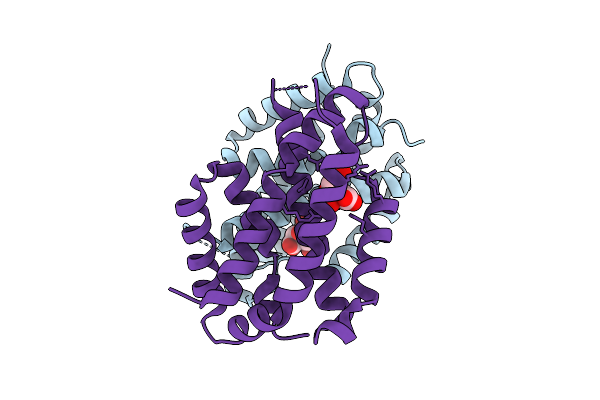 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: ISOMERASE Ligands: CIT |
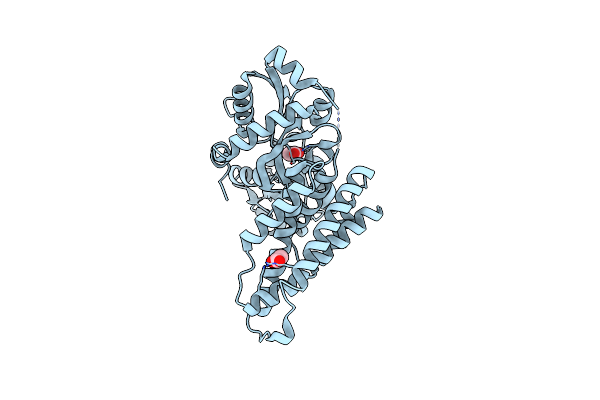 |
Bifunctional Chorismate Mutase/Cyclohexadienyl Dehydratase From Aequoribacter Fuscus
Organism: Aequoribacter fuscus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-08-30 Classification: UNKNOWN FUNCTION Ligands: ACT, CL |
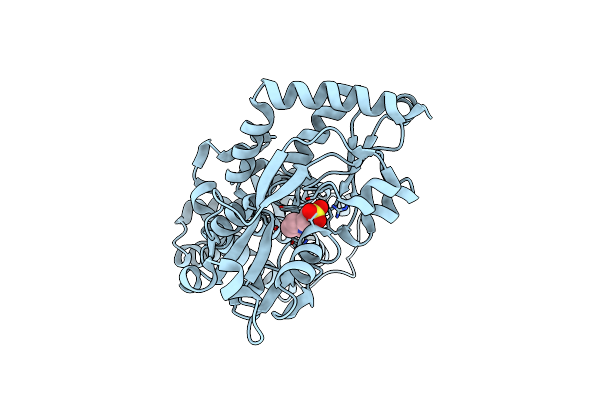 |
Bifunctional Cyclohexadienyl Dehydratase/Chorismate Mutase From Janthinobacterium Sp. Hh01
Organism: Janthinobacterium sp. hh01
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-08-30 Classification: UNKNOWN FUNCTION Ligands: MES |
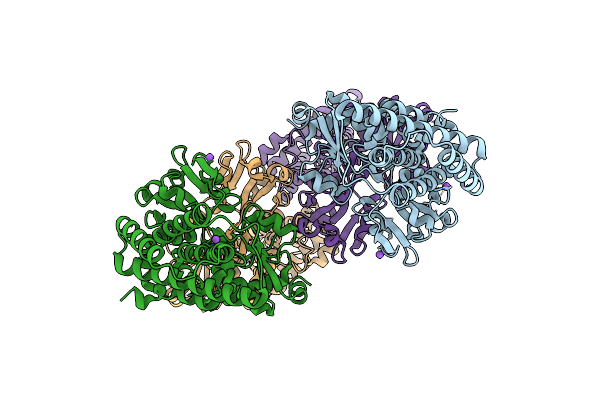 |
Bifunctional Cyclohexadienyl Dehydratase/Chorismate Mutase From Duganella Sacchari
Organism: Duganella sacchari
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2023-08-30 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of A Mycobacterium Tuberculosis Chorismate Mutase Optimized For High Autonomous Activity By Directed Evolution
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2018-08-01 Classification: ISOMERASE |
 |
High-Resolution Structure Of Chorismate Mutase From Corynebacterium Glutamicum
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2017-08-02 Classification: ISOMERASE Ligands: FMT |
 |
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2017-08-02 Classification: TRANSFERASE Ligands: MN, PEP, MPD, TRP, PO4 |
 |
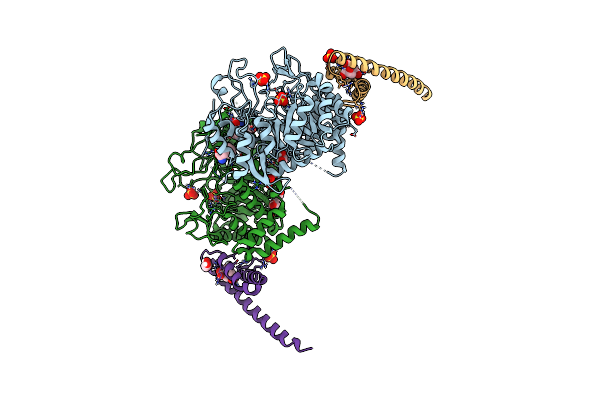
Non-Covalent Complex Of And Dahp Synthase And Chorismate Mutase From Corynebacterium Glutamicum With Bound Transition State Analog
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-08-02 Classification: TRANSFERASE/ISOMERASE Ligands: GOL, PEG, PGE, MN, PO4, TRP, PG4, TSA |
 |
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2017-08-02 Classification: SUGAR BINDING PROTEIN Ligands: MN, PEP, TRP, MPD, PGE, PO4, TRS |
 |
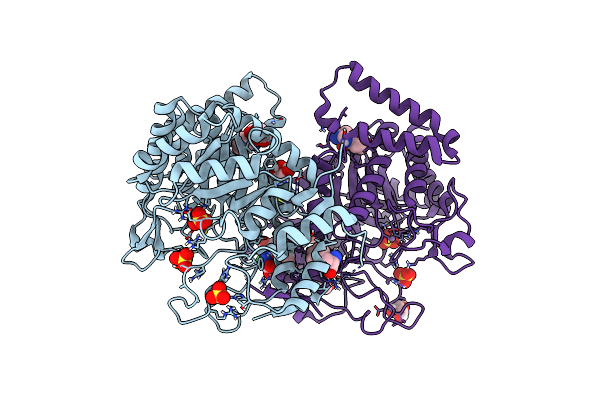
Non-Covalent Complex Of Dahp Synthase And Chorismate Mutase From Mycobacterium Tuberculosis With Bound Transition State Analog And Feedback Effectors Tyrosine And Phenylalanine
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-03-16 Classification: transferase/isomerase Ligands: MN, GOL, CL, SO4, PHE, TYR, TSA |
 |
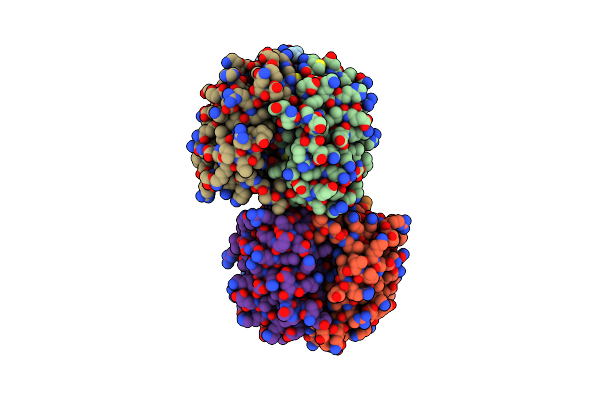
Dahp Synthase From Mycobacterium Tuberculosis, Fully Inhibited By Tyrosine, Phenylalanine, And Tryptophan
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2016-01-27 Classification: TRANSFERASE Ligands: CL, SO4, GOL, TRP, MN, PHE, TYR |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2014-04-16 Classification: ISOMERASE |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2014-04-16 Classification: ISOMERASE |
 |
Arg90Cit Chorismate Mutase Of Bacillus Subtilis In Complex With A Transition State Analog
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-04-16 Classification: ISOMERASE Ligands: TSA |
 |
Arg90Cit Chorismate Mutase Of Bacillus Subtilis In Complex With Chorismate And Prephenate
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2014-04-16 Classification: ISOMERASE Ligands: PRE, ISJ |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2012-11-07 Classification: DE NOVO PROTEIN Ligands: 3NK, MLT |
 |
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2012-11-07 Classification: LYASE Ligands: 3NK |
 |
Organism: Sulfolobus solfataricus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2012-11-07 Classification: LYASE Ligands: 3NK |
 |
Non-Covalent Complex Between Dahp Synthase And Chorismate Mutase From Mycobacterium Tuberculosis
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2009-07-07 Classification: TRANSFERASE/ISOMERASE Ligands: GOL, SO4 |
 |
Non-Covalent Complex Between Dahp Synthase And Chorismate Mutase From Mycobacterium Tuberculosis With Bound Tsa
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2009-07-07 Classification: TRANSFERASE/ISOMERASE Ligands: MN, SO4, CE1, PG4, GOL, TSA |

