Search Count: 95
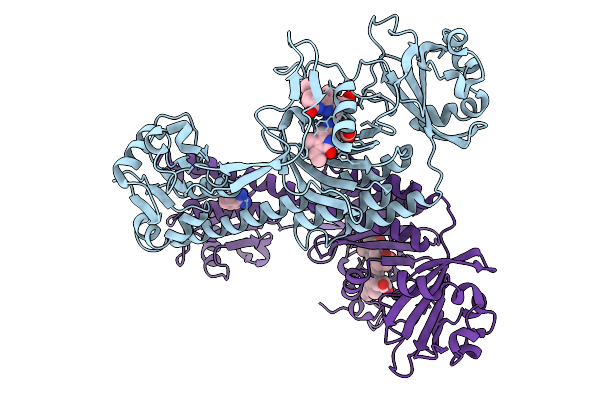 |
Photoactivation In Bacteriophytochromes, Reference (Dark) Structure For The 3 Ps Time Point
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: 3Q8, BEN |
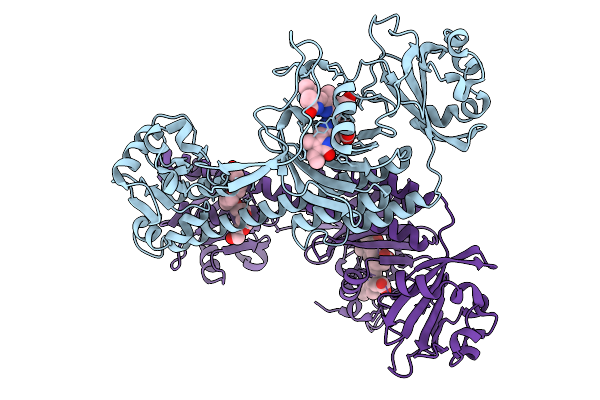 |
Photoactivation In Bacteriophytochrome, High Resolution Cryo Structure In The Dark.
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, P33 |
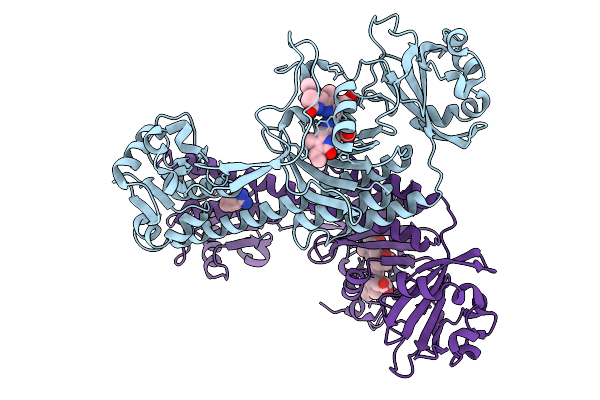 |
Photoactivation In Bacteriophytochromes, Reference (Dark) Structure For The 100 Ps Time Point
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, BEN |
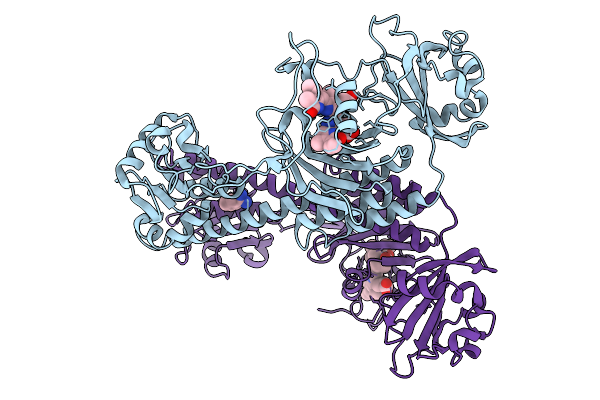 |
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: BLA, BEN |
 |
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: BLA, BEN |
 |
Crystal Structure Of The N-Terminal Domain Of Fatty Acid Kinase A (Faka) From Staphylococcus Aureus (Mn And Amp-Pnp)
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2024-11-06 Classification: LIGASE Ligands: ANP, MN |
 |
Crystal Structure Of The N-Terminal Domain Of Fatty Acid Kinase A (Faka) From Staphylococcus Aureus (Mg And Amp-Pnp)
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2024-11-06 Classification: LIGASE Ligands: ANP, MG |
 |
Crystal Structure Of The N-Terminal Domain Of Fatty Acid Kinase A (Faka) From Staphylococcus Aureus (Apo)
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-11-06 Classification: LIGASE |
 |
Crystal Structure Of The N-Terminal Domain Of Fatty Acid Kinase A (Faka) From Staphylococcus Aureus (Mg And Adp)
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2024-11-06 Classification: LIGASE Ligands: ADP, MG, PG4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2024-07-10 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: SO4, CL, GOL, YIL |
 |
Crystal Structure Of Infected Cell Protein 0 (Icp0) From Herpes Simplex Virus 1 (Proteolyzed Fragment)
Organism: Human alphaherpesvirus 1
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-02-28 Classification: PROTEIN BINDING Ligands: IOD |
 |
Crystal Structure Of Infected Cell Protein 0 (Icp0) From Herpes Simplex Virus 1 (A636-Q776)
Organism: Human alphaherpesvirus 1
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2024-02-28 Classification: PROTEIN BINDING Ligands: CL, IOD, GOL |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2024-02-07 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CL |
 |
Crystal Structure Of Root Lateral Formation Protein (Rlf) B5-Domain From Oryza Sativa
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE Ligands: HEM, SO4 |
 |
Organism: Corynebacterium diphtheriae
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-07-05 Classification: TOXIN |
 |
Organism: Corynebacterium diphtheriae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-07-05 Classification: TOXIN Ligands: APU |
 |
Structure Of The Outer-Membrane Lipoprotein Carrier Protein (Lola) From Borrelia Burgdorferi
Organism: Borreliella burgdorferi
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-03-15 Classification: LIPID TRANSPORT Ligands: PE4, CL, NA |
 |
Crystal Structure Of Apo Dps Protein (Pa0962) From Pseudomonas Aeruginosa (Orthorhombic Form)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2023-03-08 Classification: METAL BINDING PROTEIN Ligands: CL, SO4, NA |
 |
Crystal Structure Of Apo Dps Protein (Pa0962) From Pseudomonas Aeruginosa (Cubic Form)
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-03-08 Classification: METAL BINDING PROTEIN |
 |
Crystal Structure Of Iron Bound Dps Protein (Pa0962) From Pseudomonas Aeruginosa (Orthorhombic Form)
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2023-03-08 Classification: METAL BINDING PROTEIN Ligands: FE2 |

