Search Count: 125
 |
Organism: Strongylocentrotus purpuratus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: MEMBRANE PROTEIN |
 |
Organism: Salpingoeca rosetta
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: MEMBRANE PROTEIN |
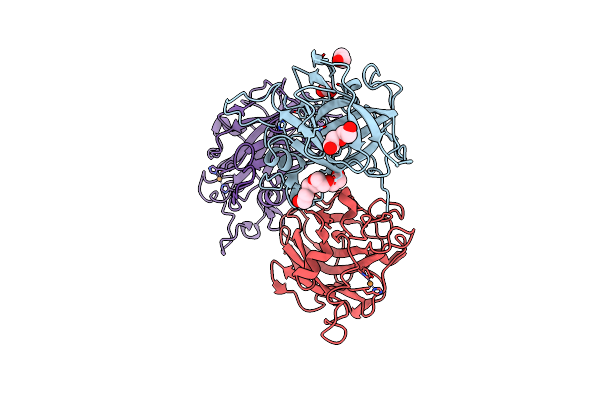 |
Crystal Structure Of The Catalytic Domain Of An Aa9 Lytic Polysaccharide Monooxygenase From Thermothelomyces Thermophilus (Ttlpmo9F)
Organism: Thermothelomyces thermophilus atcc 42464
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2024-12-25 Classification: METAL BINDING PROTEIN Ligands: PEG, PG4, EDO, FMT, CU |
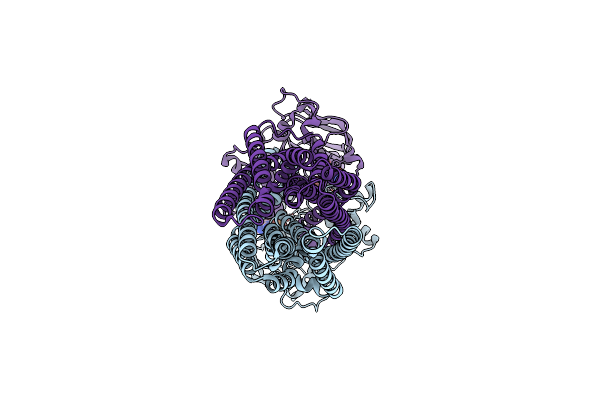 |
Cryo-Em Structure Of The Abc Transporter Pcat1 Cysteine-Free Core Bound With Mgatp And Vi
Organism: Acetivibrio thermocellus atcc 27405
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: TRANSPORT PROTEIN Ligands: ATP, MG, ADP, VO4 |
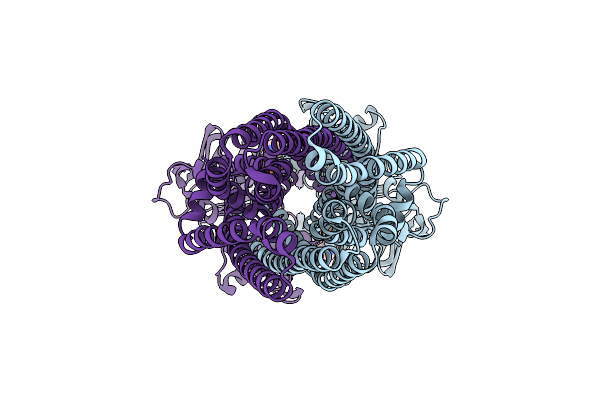 |
Cryo-Em Structure Of The Abc Transporter Pcat1 Cysteine-Free Core Bound With Mgadp
Organism: Acetivibrio thermocellus atcc 27405
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: TRANSPORT PROTEIN Ligands: ADP, MG |
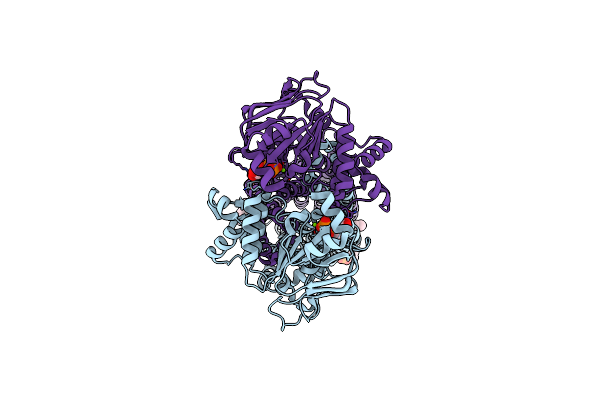 |
Cryo-Em Structure Of The Abc Transporter Pcat1 Cysteine-Free Core Bound With Mgadp And Substrate
Organism: Acetivibrio thermocellus atcc 27405
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: TRANSPORT PROTEIN Ligands: ADP, MG, A1ACX, POV |
 |
Cryo-Em Structure Of The Abc Transporter Pcat1 Cysteine-Free Core Bound With Atp
Organism: Acetivibrio thermocellus atcc 27405
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: TRANSPORT PROTEIN Ligands: ATP |
 |
Cryo-Em Structure Of The Abc Transporter Pcat1 Bound With Mgadp And Substrate
Organism: Acetivibrio thermocellus atcc 27405
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: TRANSPORT PROTEIN Ligands: ADP, MG, A1ACX, POV |
 |
Cryo-Em Structure Of The Abc Transporter Pcat1 Bound With Adp And Substrate
Organism: Acetivibrio thermocellus atcc 27405
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: TRANSPORT PROTEIN Ligands: ADP, A1ACX, POV |
 |
Organism: Acetivibrio thermocellus atcc 27405
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: TRANSPORT PROTEIN |
 |
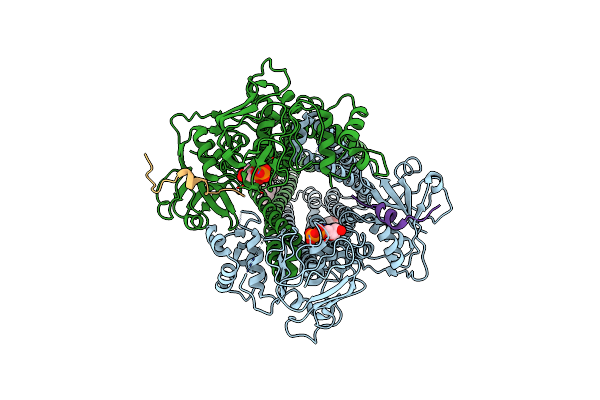
Cryo-Em Structure Of The Abc Transporter Pcat1 Bound With Atp And Substrate
Organism: Acetivibrio thermocellus atcc 27405
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: TRANSPORT PROTEIN Ligands: ATP |
 |
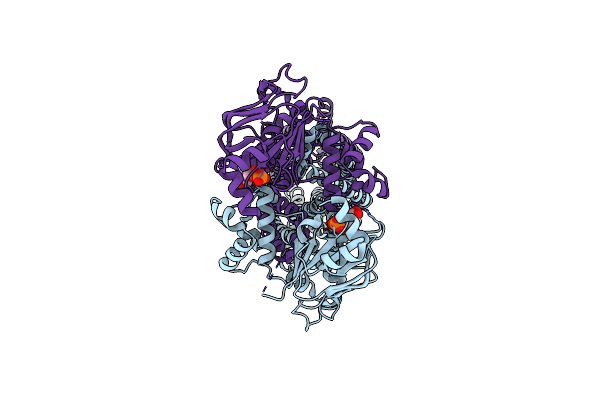
Cryo-Em Structure Of The Cysteine-Free Abc Transporter Pcat1 Bound With Adp And Substrate
Organism: Acetivibrio thermocellus atcc 27405
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: TRANSPORT PROTEIN Ligands: ADP, A1ACX |
 |
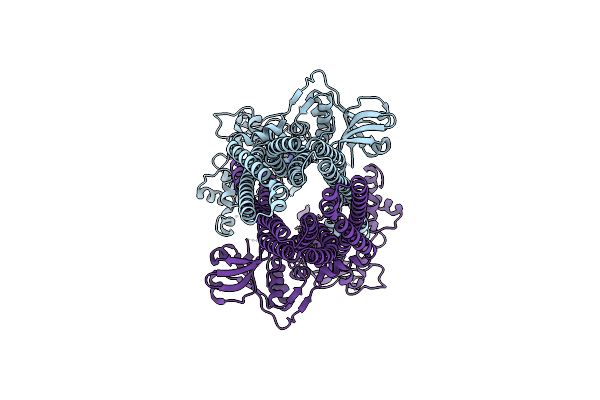
Organism: Acetivibrio thermocellus atcc 27405
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: TRANSPORT PROTEIN Ligands: ADP |
 |
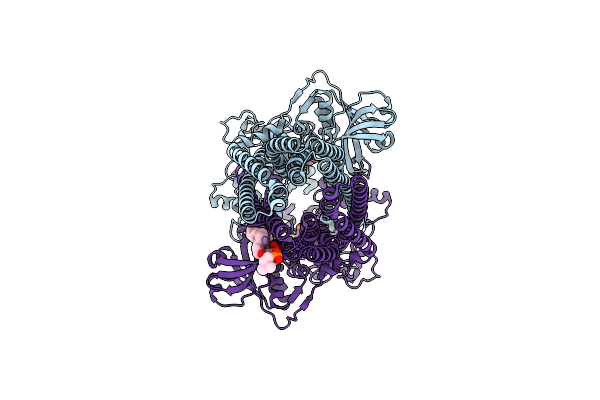
Organism: Acetivibrio thermocellus atcc 27405
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: TRANSPORT PROTEIN Ligands: ADP, MG |
 |
Organism: Acetivibrio thermocellus atcc 27405
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: TRANSPORT PROTEIN Ligands: ADP, MG, POV |
 |
Organism: Acetivibrio thermocellus atcc 27405
Method: ELECTRON MICROSCOPY Release Date: 2024-10-30 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of Thermothelomyces Thermophila Gh30 (Double Mutant Ee) In Complex With Xylopentaose
Organism: Thermothelomyces
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-05-22 Classification: HYDROLASE Ligands: EDO, F |
 |
Crystal Structure Of Thermothelomyces Thermophila Gh30 (Double Mutant Ee) In Complex With Xylotriose.
Organism: Thermothelomyces
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-05-22 Classification: HYDROLASE Ligands: EDO, PEG |
 |
Crystal Structure Of Thermothelomyces Thermophila (Double Mutant Ee) In Complex With Aldotetrauronic Acid
Organism: Thermothelomyces thermophilus
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2024-05-22 Classification: HYDROLASE Ligands: EDO |
 |
Organism: Thermothelomyces thermophilus
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2024-03-20 Classification: HYDROLASE Ligands: B3P, PEG, ACT, EDO, NAG, PG4 |

