Search Count: 24
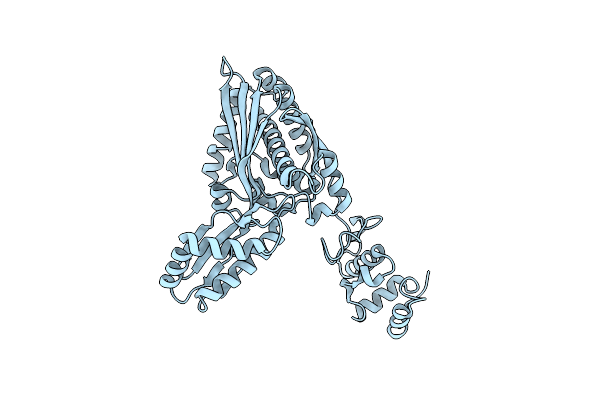 |
Cryoem Structure Of Apo Form Of Catalytic Domain Of Human Hmg-Coa Reductase
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.06 Å Release Date: 2025-02-26 Classification: OXIDOREDUCTASE |
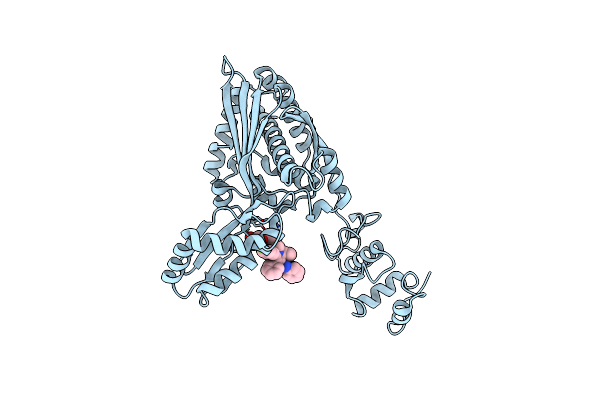 |
Cryoem Structure Of Catalytic Domain Of Human Hmg-Coa Reductase With Its Inhibitor Atorvastatin
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.26 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: 117 |
 |
Bat Influenza A Polymerase Pre-Termination Complex With Pyrophosphate Using 44-Mer Vrna Template With Mutated Oligo(U) Sequence
Organism: Influenza a virus (a/little yellow-shouldered bat/guatemala/060/2010(h17n10)), Influenza b virus
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: VIRAL PROTEIN Ligands: M4H, MG, POP |
 |
Bat Influenza A Polymerase Elongation Complex With Incoming Utp Analogue (Core + Endonuclease Only)
Organism: Influenza a virus (a/little yellow-shouldered bat/guatemala/060/2010(h17n10)), Influenza b virus
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: VIRAL PROTEIN Ligands: M4H, MG, 2KH |
 |
Organism: Influenza a virus (a/little yellow-shouldered bat/guatemala/060/2010(h17n10)), Influenza b virus
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: VIRAL PROTEIN Ligands: MG |
 |
Bat Influenza A Polymerase Product Dissociation Complex Using 44-Mer Vrna Template With Mutated Oligo(U) Sequence
Organism: Influenza a virus (a/little yellow-shouldered bat/guatemala/060/2010(h17n10)), Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: VIRAL PROTEIN |
 |
Bat Influenza A Polymerase Stuttering Complex Using 44-Mer Vrna Template With Intact Oligo(U) Sequence
Organism: Influenza a virus (a/little yellow-shouldered bat/guatemala/060/2010(h17n10)), Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: VIRAL PROTEIN Ligands: MG |
 |
Bat Influenza A Polymerase Product Dissociation Complex Using 44-Mer Vrna Template With Intact Oligo(U) Sequence
Organism: Influenza a virus (a/little yellow-shouldered bat/guatemala/060/2010(h17n10)), Influenza b virus
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: VIRAL PROTEIN |
 |
Bat Influenza A Polymerase Elongation Complex With Incoming Utp Analogue (Complete Polymerase)
Organism: Influenza a virus (a/little yellow-shouldered bat/guatemala/060/2010(h17n10))
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: VIRAL PROTEIN Ligands: M4H, MG, 2KH |
 |
Organism: Influenza b virus
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: VIRAL PROTEIN |
 |
Organism: Influenza a virus (a/little yellow-shouldered bat/guatemala/060/2010(h17n10)), Influenza b virus
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: VIRAL PROTEIN Ligands: MG |
 |
Organism: Influenza a virus (a/zhejiang/dtid-zju01/2013(h7n9))
Method: X-RAY DIFFRACTION Resolution:3.33 Å Release Date: 2020-04-15 Classification: VIRAL PROTEIN Ligands: MG |
 |
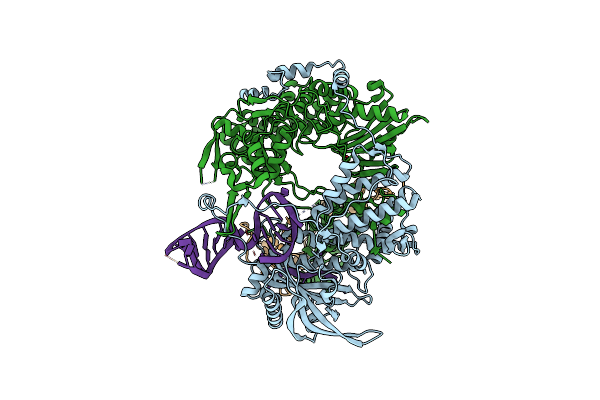
Bat Influenza A Polymerase Termination Complex With Pyrophosphate Using 44-Mer Vrna Template With Mutated Oligo(U) Sequence
Organism: Influenza a virus (a/little yellow-shouldered bat/guatemala/060/2010(h17n10)), Influenza b virus
Method: ELECTRON MICROSCOPY Release Date: 2020-04-15 Classification: VIRAL PROTEIN Ligands: M4H, MG, POP |
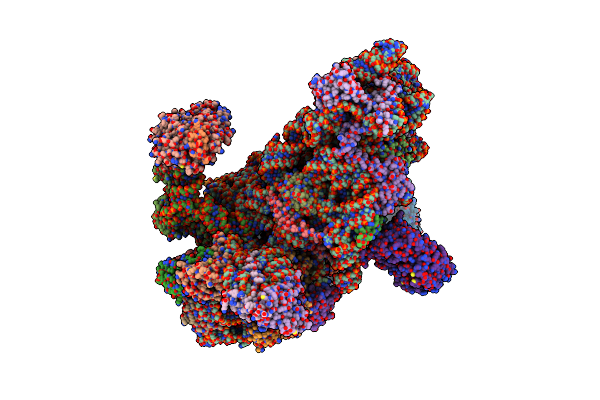 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:6.30 Å Release Date: 2018-03-14 Classification: RIBOSOME |
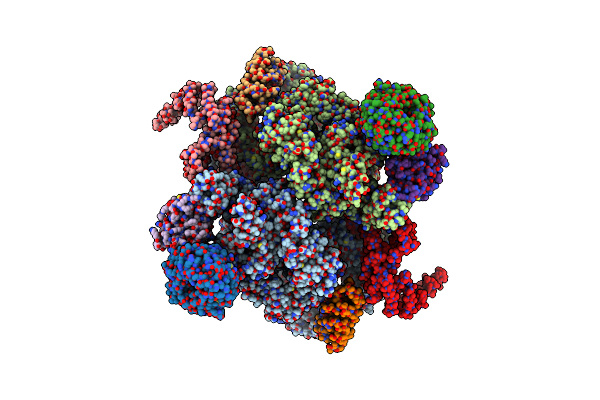 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Target Of Rapamycin Complex 2
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: ELECTRON MICROSCOPY Resolution:7.90 Å Release Date: 2017-12-06 Classification: SIGNALING PROTEIN |
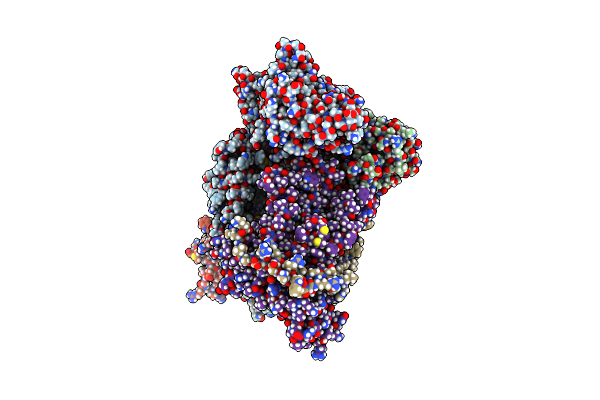 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2016-12-28 Classification: CHAPERONE |
 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Resolution:5.70 Å Release Date: 2015-03-25 Classification: RIBOSOME Ligands: MG |
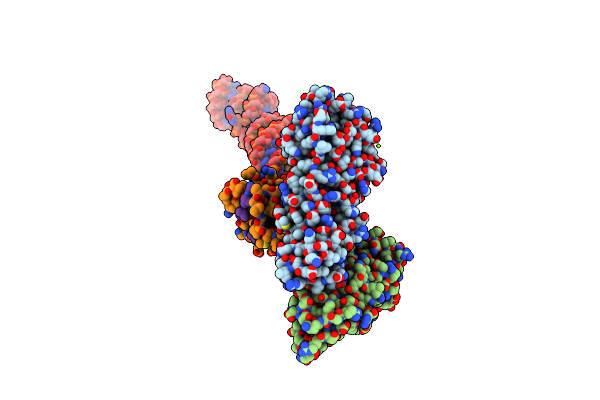 |
Structural Basis Of Signal Sequence Surveillance And Selection By The Srp-Sr Complex
Organism: Escherichia coli, Sulfolobus solfataricus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Resolution:12.00 Å Release Date: 2013-03-06 Classification: PROTEIN TRANSPORT Ligands: MG |
 |
Organism: Artificial gene, Ovis aries
Method: X-RAY DIFFRACTION Resolution:4.17 Å Release Date: 2012-07-18 Classification: CELL CYCLE Ligands: GTP, MG, GDP |
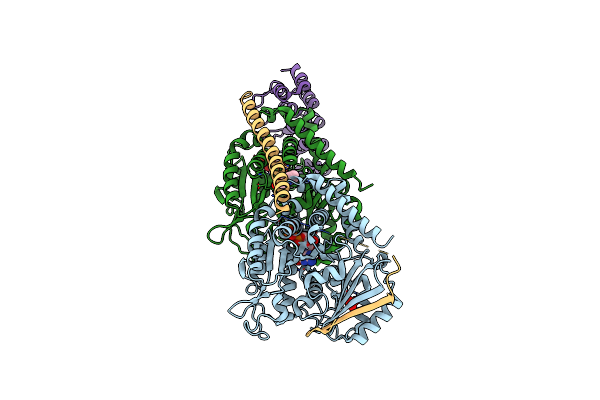 |
Organism: Artificial gene, Ovis aries
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2012-07-18 Classification: CELL CYCLE Ligands: GTP, MG, SO4, GDP, MES |

