Search Count: 49
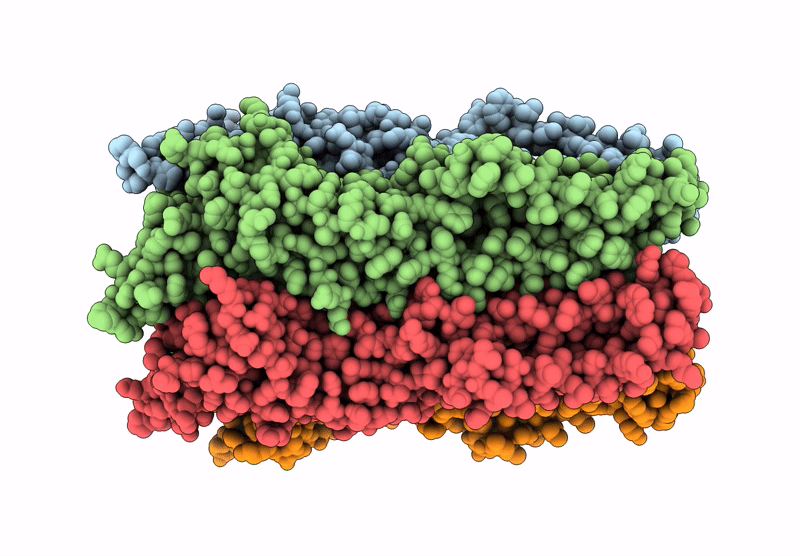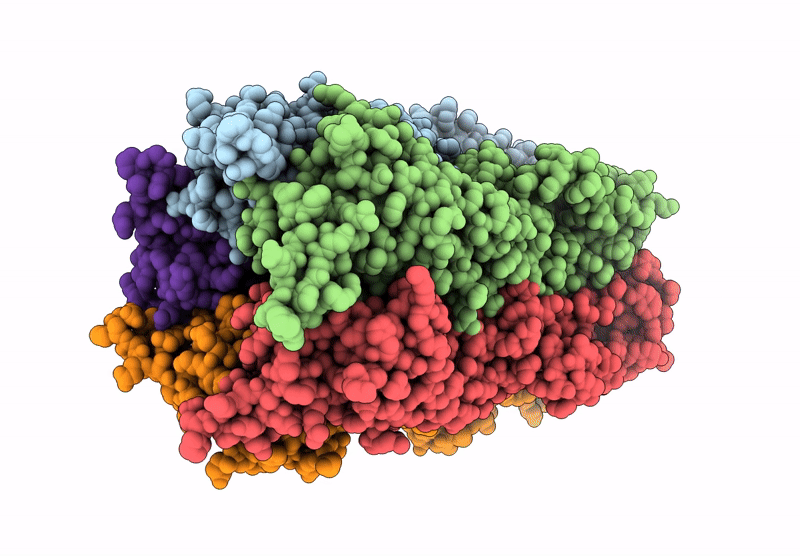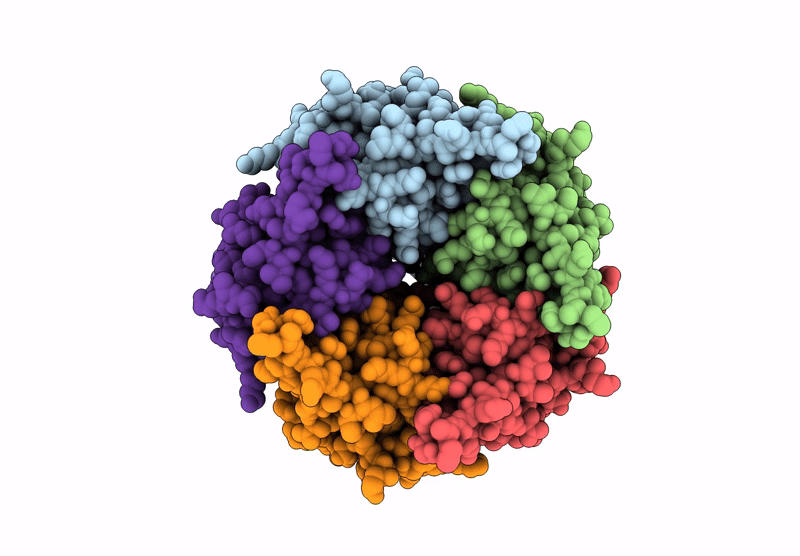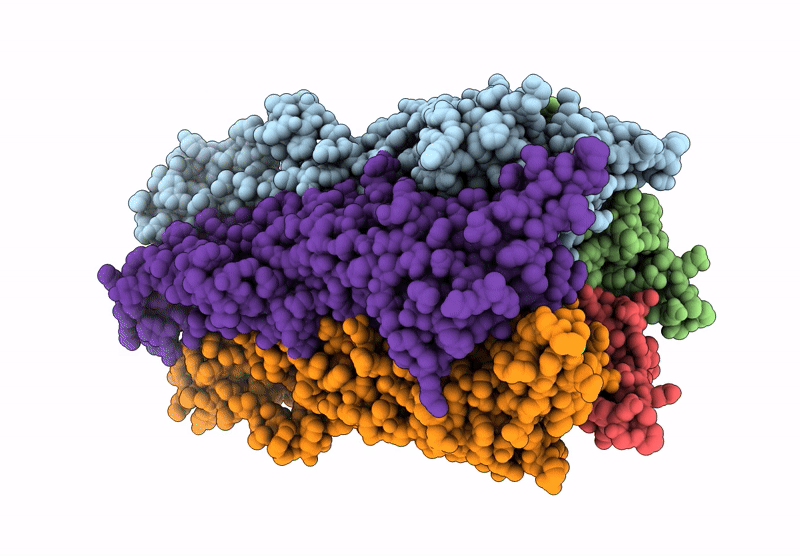 |
The Stelic Pentameric Ligand-Gated Ion Channel (Wild-Type) In Complex With 4-Bromophenethylamine
Organism: Endosymbiont of tevnia jerichonana (vent tica)
Method: X-RAY DIFFRACTION Resolution:3.01 Å Release Date: 2025-03-26 Classification: MEMBRANE PROTEIN Ligands: BNG, VMT |
 |
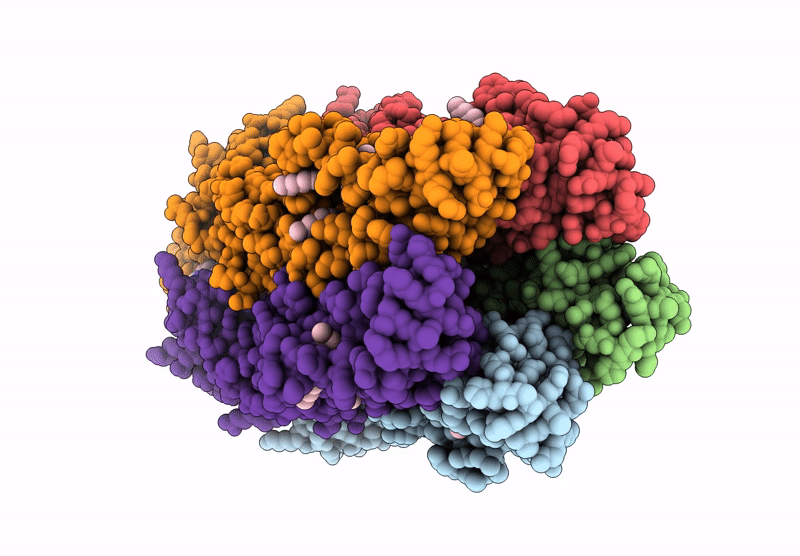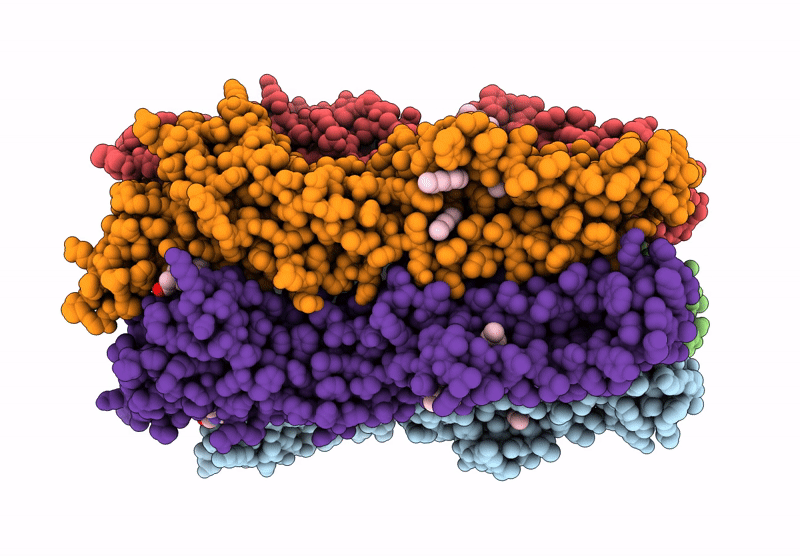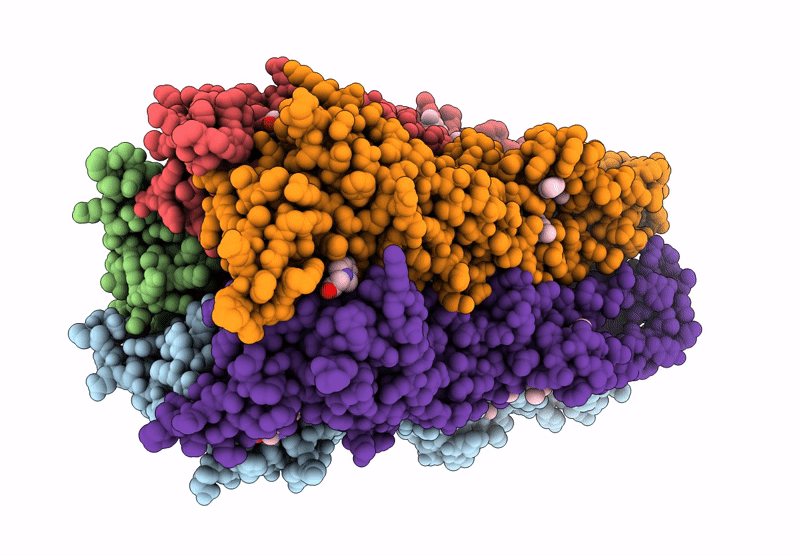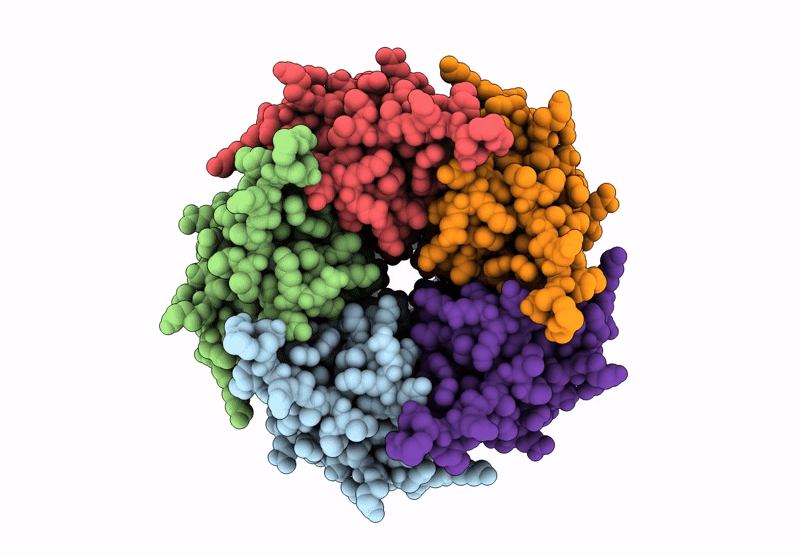
The Stelic Pentameric Ligand-Gated Ion Channel (Wild-Type) In Complex With 4-Bromoamphetamine
Organism: Endosymbiont of tevnia jerichonana (vent tica)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2025-03-26 Classification: MEMBRANE PROTEIN Ligands: BNG, A1H7R |
 |
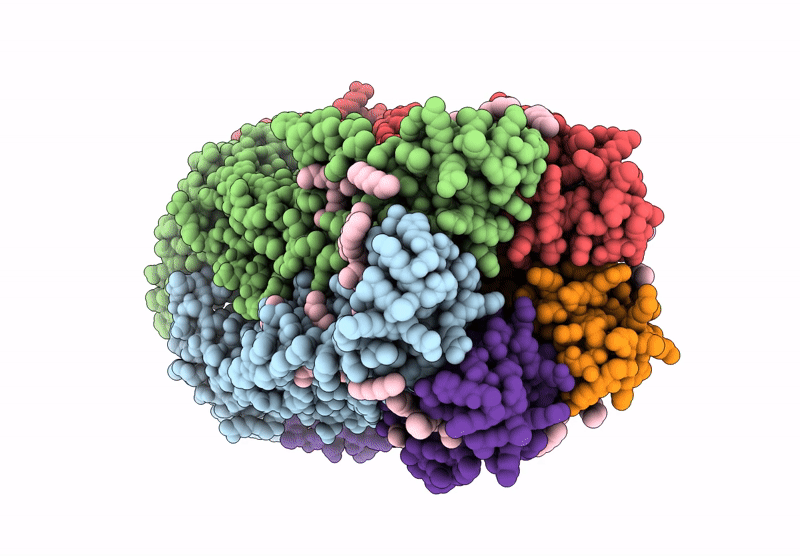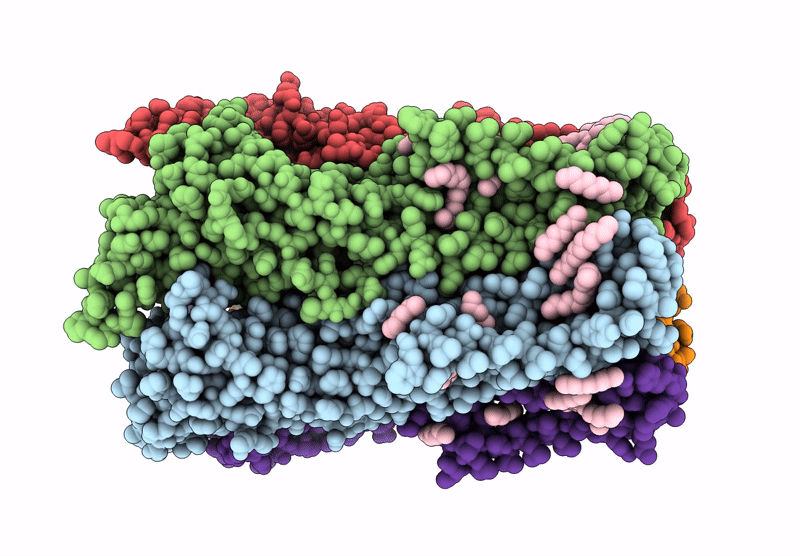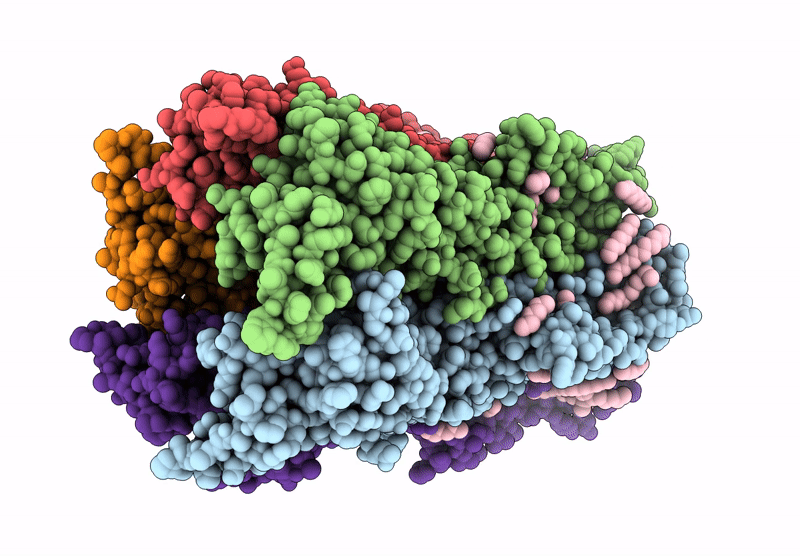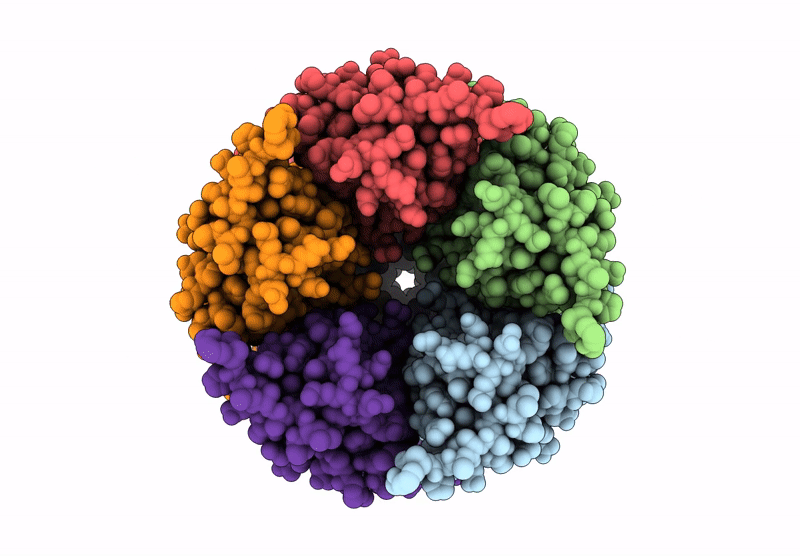
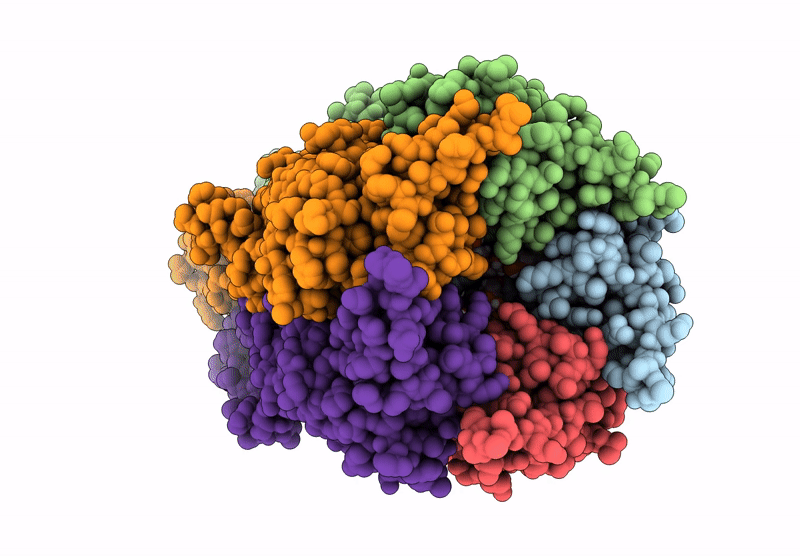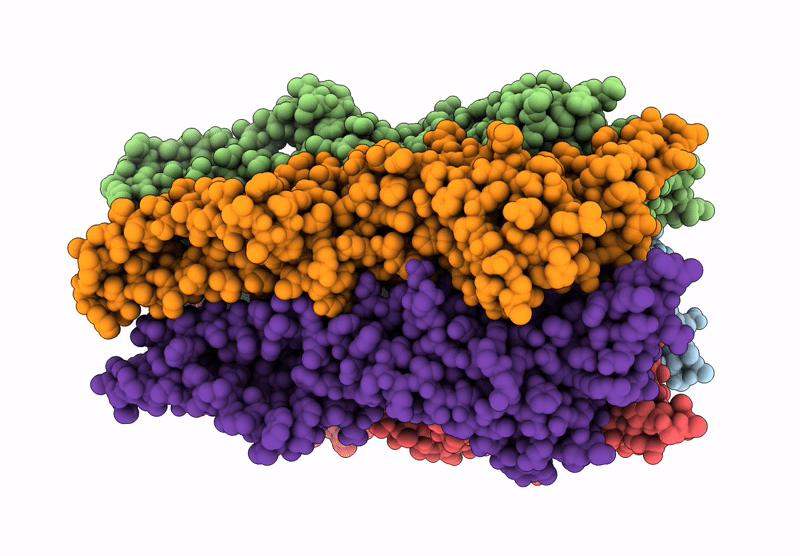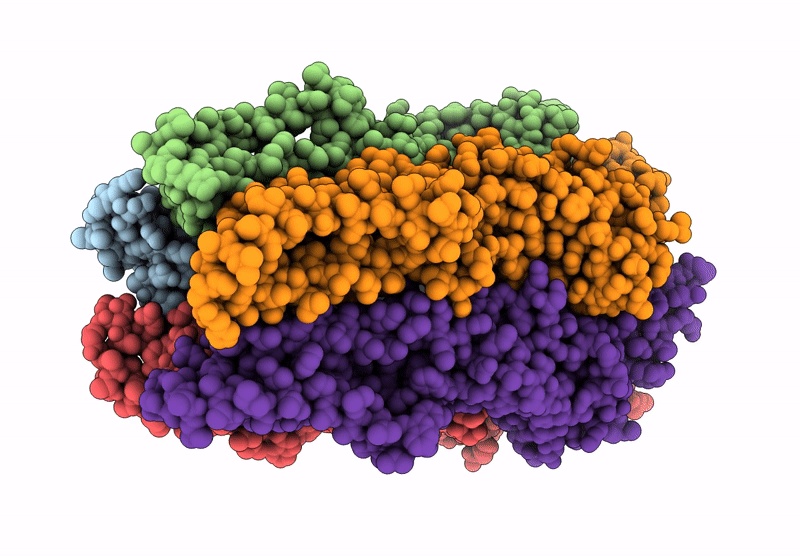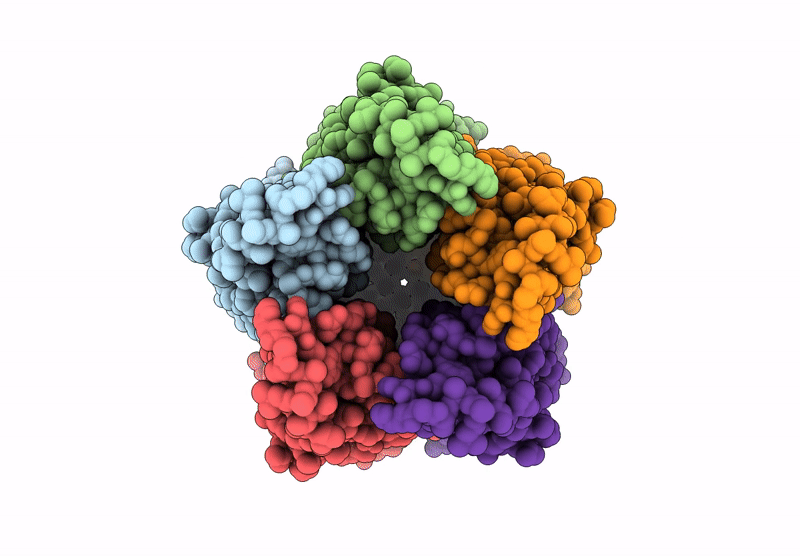
Cryoem Structure Of Stelic Nanodisc In Complex With Fluorinated Fos-Choline-8 In Open State
Organism: Endosymbiont of tevnia jerichonana (vent tica)
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: TRANSLOCASE Ligands: A1H8K, HEX, EPE |
 |
Organism: Endosymbiont of tevnia jerichonana (vent tica)
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: TRANSLOCASE Ligands: D12, OCT, C14, HEX, D10 |
 |
Cryoem Structure Of Open Stelic In Detergent, In Complex With N-Dodecyl-Beta-Maltoside
Organism: Endosymbiont of tevnia jerichonana (vent tica)
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: MEMBRANE PROTEIN Ligands: LMT |
 |
Organism: Endosymbiont of tevnia jerichonana (vent tica)
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: MEMBRANE PROTEIN |
 |
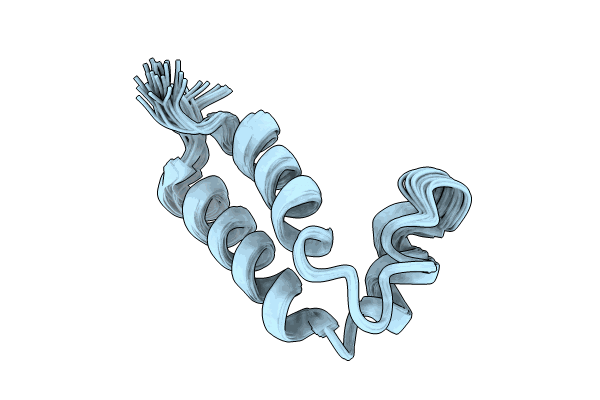
The Molecular Structure Of A Beta-1,4-D-Xylosidase From The Probiotic Bacterium Levilactobacillus Brevis
Organism: Levilactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-01-15 Classification: CARBOHYDRATE |
 |
Organism: Apilactobacillus kunkeei
Method: SOLUTION NMR Release Date: 2024-02-21 Classification: UNKNOWN FUNCTION |
 |
Organism: Apilactobacillus kunkeei
Method: SOLUTION NMR Release Date: 2024-02-21 Classification: UNKNOWN FUNCTION |
 |
Organism: Apilactobacillus kunkeei
Method: SOLUTION NMR Release Date: 2024-02-21 Classification: UNKNOWN FUNCTION |
 |
Sialidases And Fucosidases Of Akkermansia Muciniphila Are Key For Rapid Growth On Colonic Mucin And Nutrient Sharing Amongst Mucin-Associated Human Gut Microbiota
Organism: Akkermansia muciniphila
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2023-03-01 Classification: HYDROLASE Ligands: DAN, GAL, EDO, CL, CA |
 |
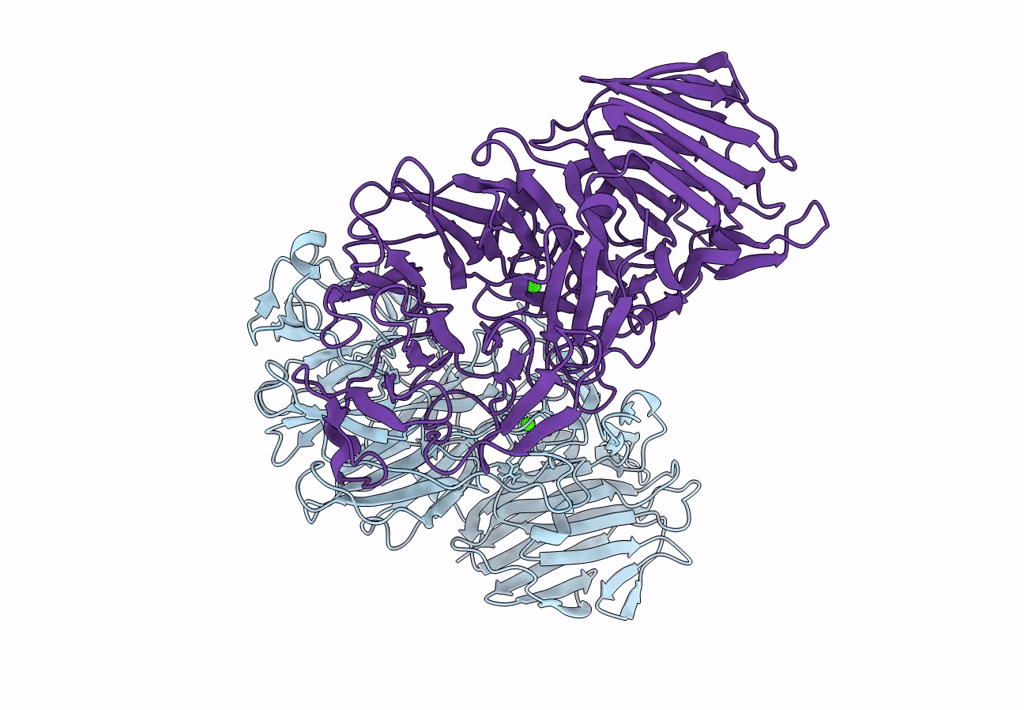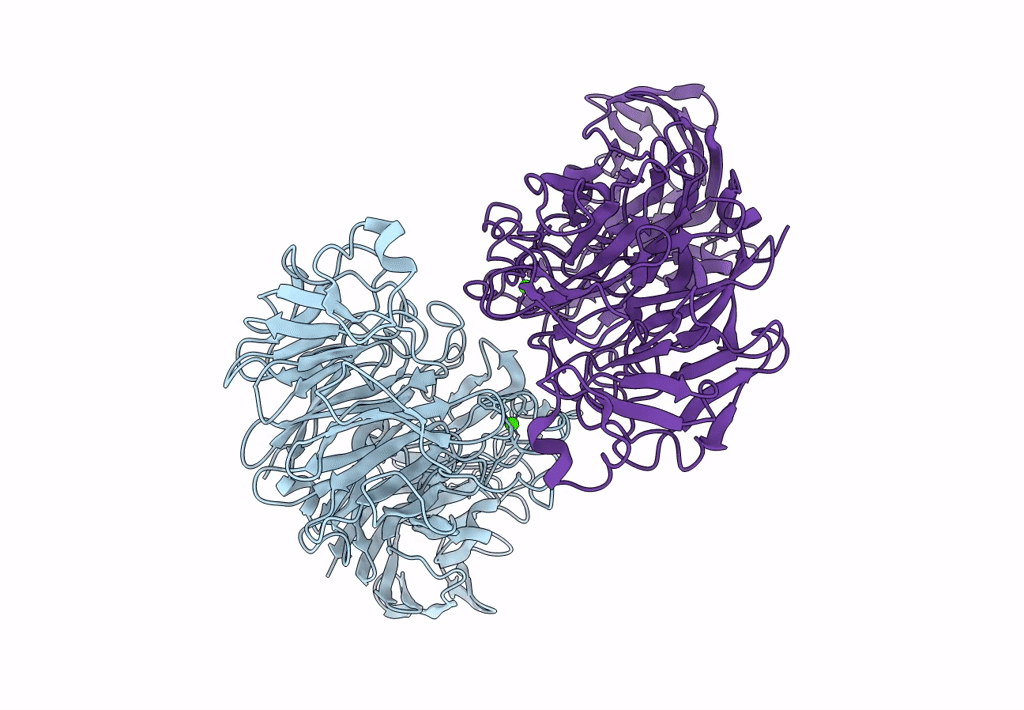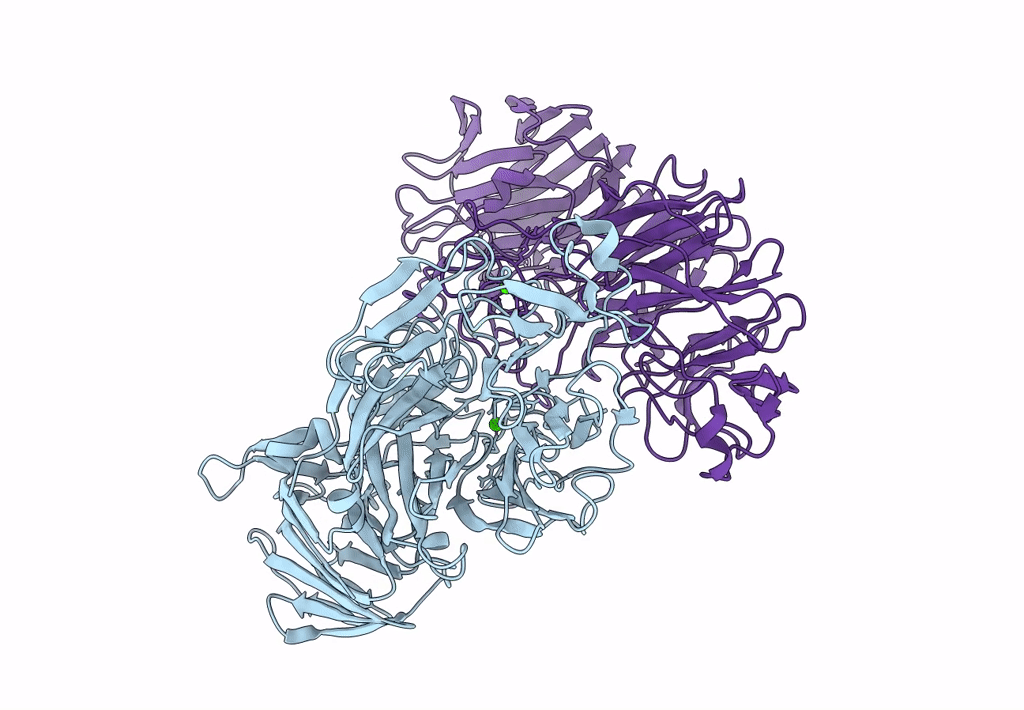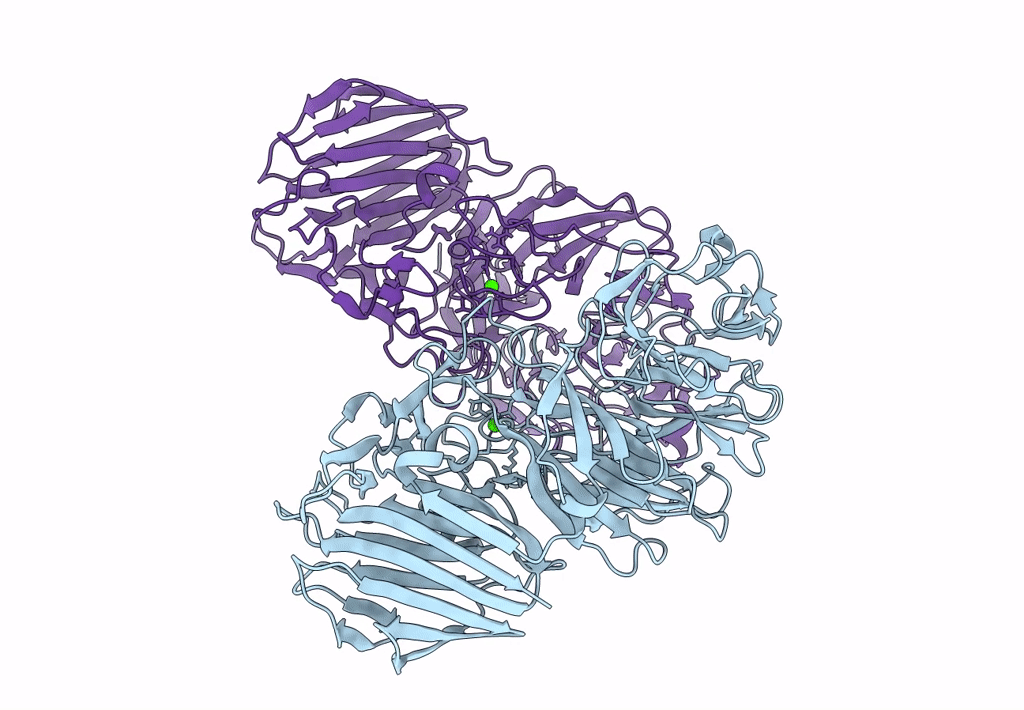
Sialidases And Fucosidases Of Akkermansia Muciniphila Are Key For Rapid Growth On Colonic Mucin And Nutrient Sharing Amongst Mucin-Associated Human Gut Microbiota
Organism: Akkermansia muciniphila
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2023-03-01 Classification: HYDROLASE Ligands: SLB, IMD, EDO, CL, CA, SO4, DAN |
 |
Sialidases And Fucosidases Of Akkermansia Muciniphila Are Key For Rapid Growth On Colonic Mucin And Nutrient Sharing Amongst Mucin-Associated Human Gut Microbiota
Organism: Akkermansia muciniphila
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2023-03-01 Classification: HYDROLASE Ligands: CA, CL |
 |
Sialidases And Fucosidases Of Akkermansia Muciniphila Are Key For Rapid Growth On Colonic Mucin And Nutrient Sharing Amongst Mucin-Associated Human Gut Microbiota
Organism: Akkermansia muciniphila
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-03-01 Classification: HYDROLASE Ligands: CA |
 |
Organism: Thermus phage g20c
Method: X-RAY DIFFRACTION Resolution:2.97 Å Release Date: 2022-11-02 Classification: VIRAL PROTEIN |
 |
Organism: Thermus phage tsp4
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2022-11-02 Classification: VIRAL PROTEIN Ligands: TMP, MG, TB, CL |
 |
Organism: Thermus parvatiensis
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2022-06-22 Classification: HYDROLASE Ligands: ZN, GOL, EPE, SO4, NA, CL |
 |
Structure And Folding Of A 600-Million-Year-Old Nuclear Coactivator Binding Domain Suggest Conservation Of Dynamic Properties
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2022-04-20 Classification: DNA BINDING PROTEIN |
 |
Structure And Folding Of A 600-Million-Year-Old Nuclear Coactivator Binding Domain Suggest Conservation Of Dynamic Properties
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2022-04-20 Classification: DNA BINDING PROTEIN |
 |
Organism: Sulfolobales beppu filamentous virus 3
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-03-02 Classification: VIRAL PROTEIN Ligands: GOL, NI |

