Search Count: 16
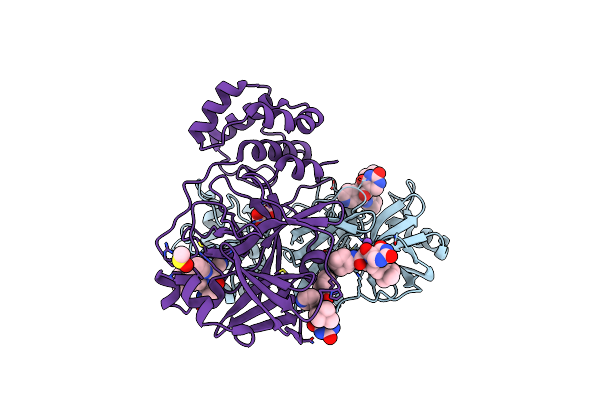 |
Co-Crystal Structure Of The Sars-Cov2 Main Protease Nsp5 With An Uracil-Carrying X77-Like Inhibitor
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-01-31 Classification: ANTIVIRAL PROTEIN Ligands: MLI, YQN, NA, DMS |
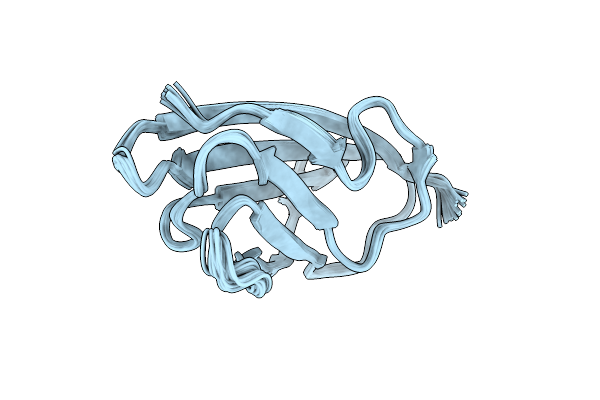 |
Organism: Pseudomonas fluorescens
Method: SOLUTION NMR Release Date: 2021-01-13 Classification: CHAPERONE |
 |
Structure Of The N-Terminal Domain Of The Metalloprotease Prtv From Vibrio Cholerae
|
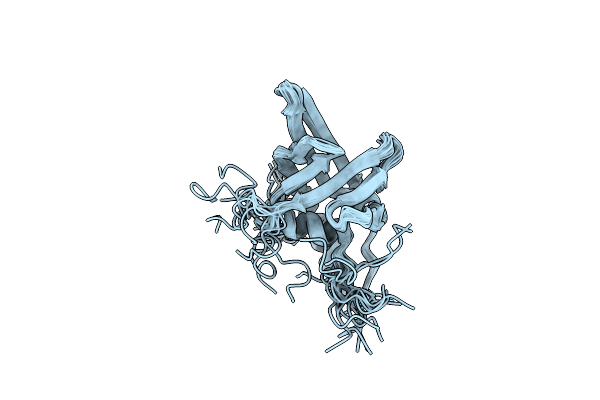 |
Organism: Pseudomonas aeruginosa
Method: SOLUTION NMR Release Date: 2014-04-30 Classification: STRUCTURAL PROTEIN |
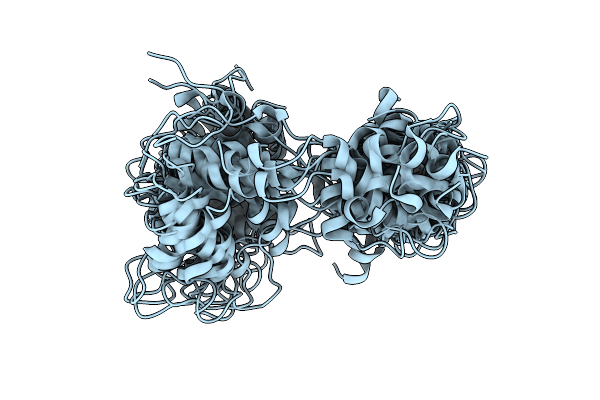 |
Organism: Entamoeba histolytica
Method: SOLUTION NMR Release Date: 2013-06-05 Classification: STRUCTURAL PROTEIN |
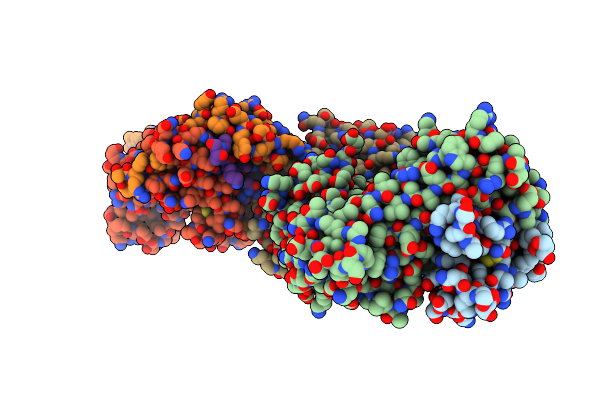 |
Crystal Structure Of The Ternary Complex Between Human T Cell Receptor, Staphylococcal Enterotoxin H And Human Major Histocompatibility Complex Class Ii
Organism: Homo sapiens, Staphylococcus aureus, Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-11-24 Classification: IMMUNE SYSTEM Ligands: GOL, NA |
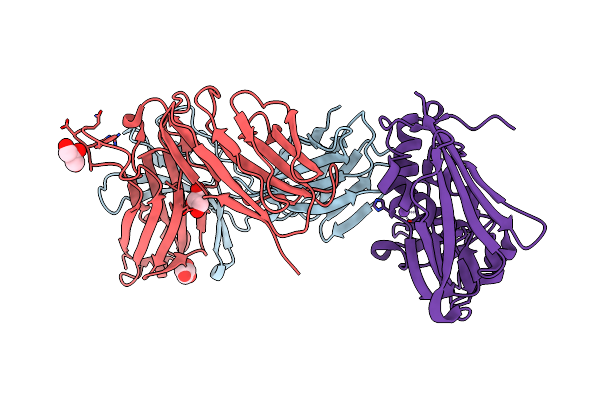 |
Crystal Structure Of The Complex Between Human T Cell Receptor And Staphylococcal Enterotoxin
Organism: Homo sapiens, Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-11-24 Classification: IMMUNE SYSTEM Ligands: GOL, NA |
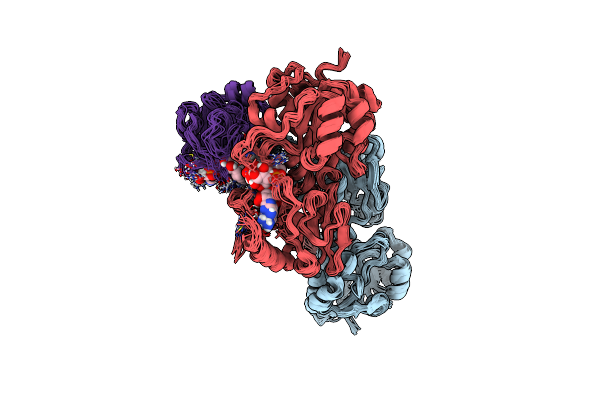 |
Complex Of The Domain I And Domain Iii Of Escherichia Coli Transhydrogenase
Organism: Escherichia coli
Method: SOLUTION NMR Release Date: 2006-08-29 Classification: OXIDOREDUCTASE Ligands: NAD, NAP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2005-09-13 Classification: OXIDOREDUCTASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2005-09-13 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2005-09-13 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Crystal Structure Of The Double Mutant Cys3Ser/Ser100Pro From Pseudomonas Aeruginosa At 1.8 A Resolution
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2002-05-16 Classification: ELECTRON TRANSPORT Ligands: CU |
 |
Crystal Structure Of The Disulphide Bond-Deficient Azurin Mutant C3A/C26A: How Important Is The S-S Bond For Folding And Stability?
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2000-08-09 Classification: ELECTRON TRANSPORT Ligands: CU |
 |
The Complex Of Cytochrome F And Plastocyanin Determined With Paramagnetic Nmr. Based On The Structures Of Cytochrome F And Plastocyanin, 10 Structures
Organism: Spinacia oleracea, Brassica rapa
Method: SOLUTION NMR Release Date: 1998-04-08 Classification: COMPLEX (ELECTRON TRANSPORT PROTEINS) Ligands: CU, HEC |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 1997-04-21 Classification: ELECTRON TRANSPORT Ligands: CU |
 |
Crystal Structure Of The Azurin Mutant Nickel-Trp48Met From Pseudomonas Aeruginosa At 2.2 Angstroms Resolution
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 1995-02-27 Classification: ELECTRON TRANSPORT (COPPER) Ligands: NI, NO3 |

