Search Count: 17
 |
Organism: Illicium anisatum
Method: SOLUTION NMR Release Date: 2022-11-09 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Zika virus (strain mr 766)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-03-29 Classification: TRANSFERASE Ligands: SAH, ZN, SO4 |
 |
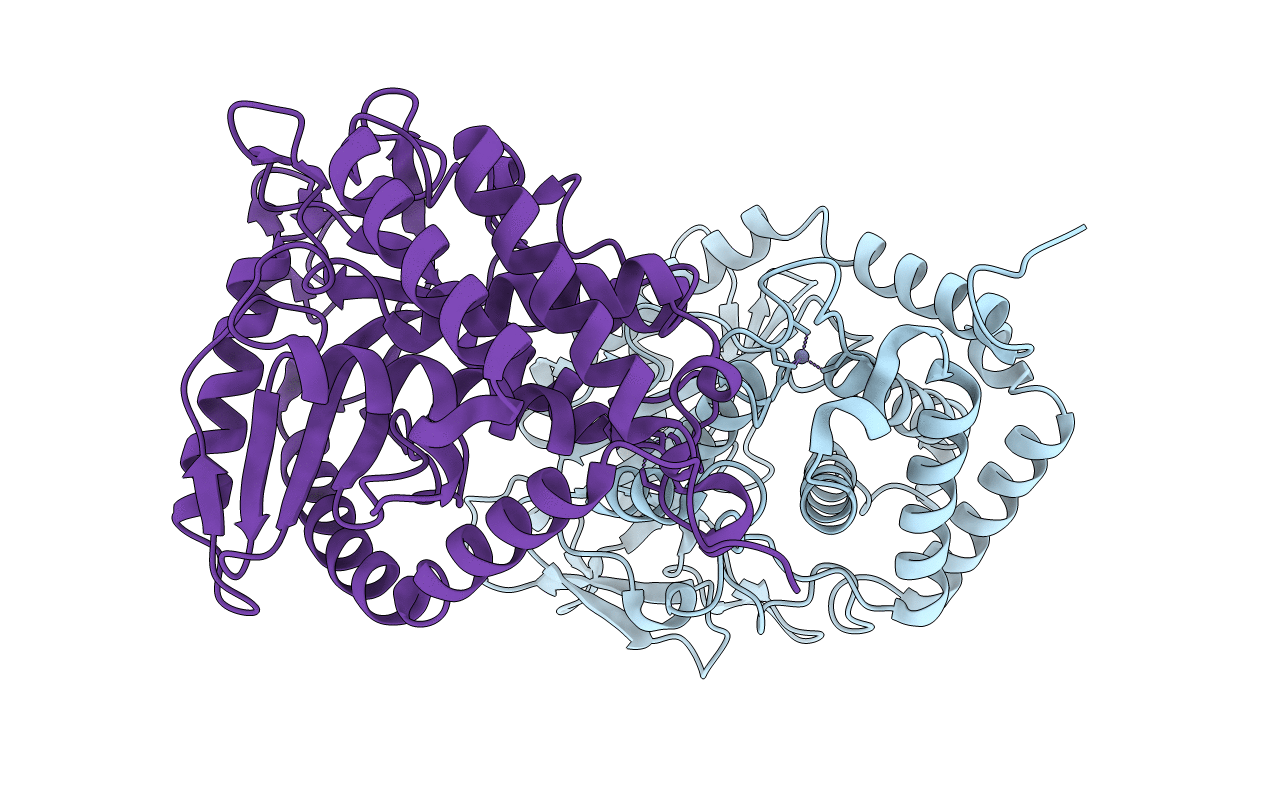
Organism: Zika virus (strain mr 766)
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2017-03-29 Classification: TRANSFERASE Ligands: ZN |
 |
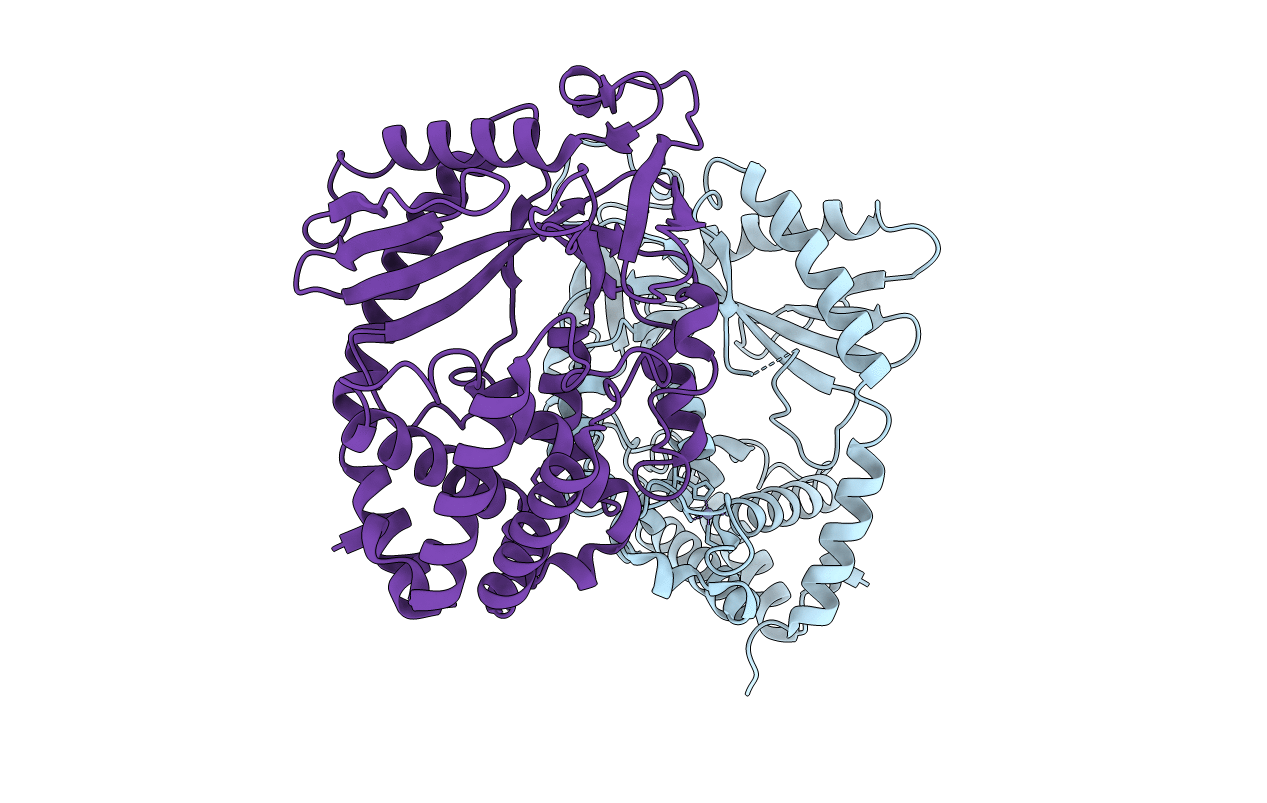
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2013-12-25 Classification: TRANSFERASE Ligands: ZN |
 |
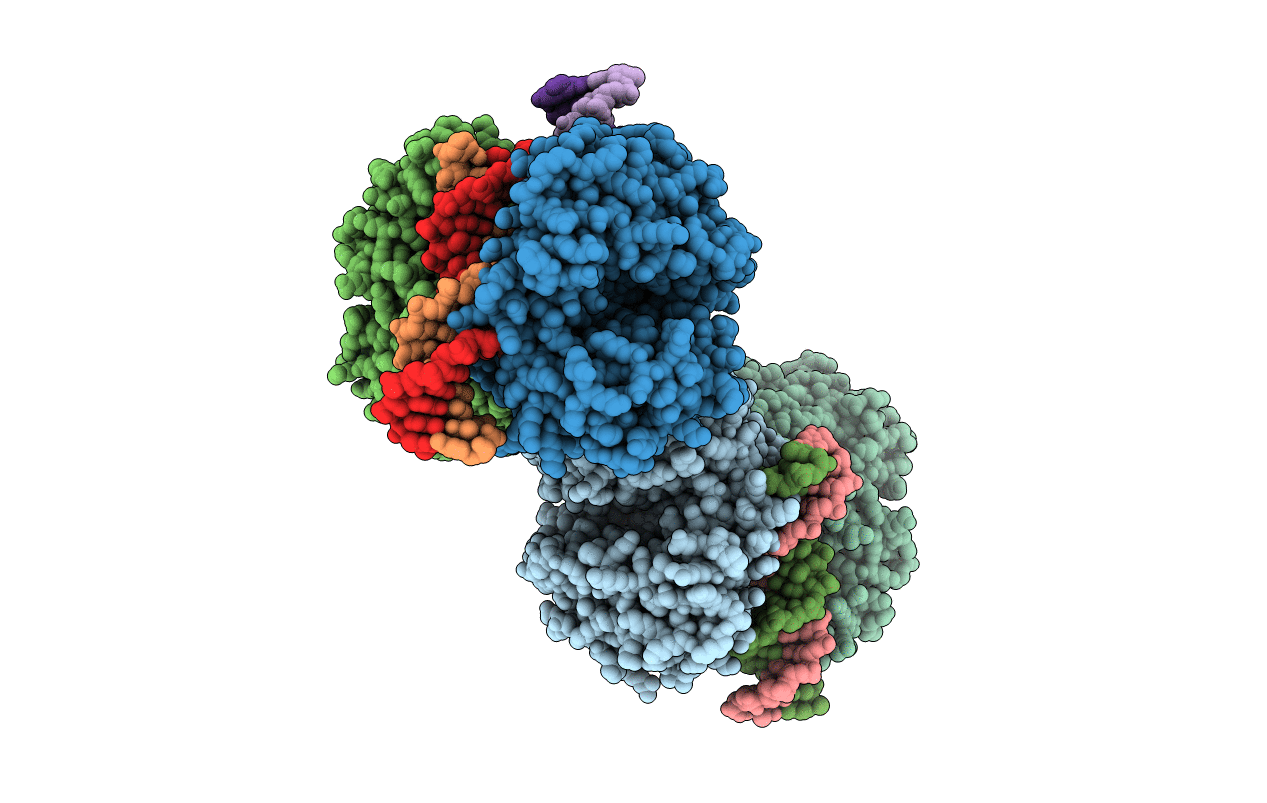
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2013-12-25 Classification: TRANSFERASE Ligands: ZN |
 |
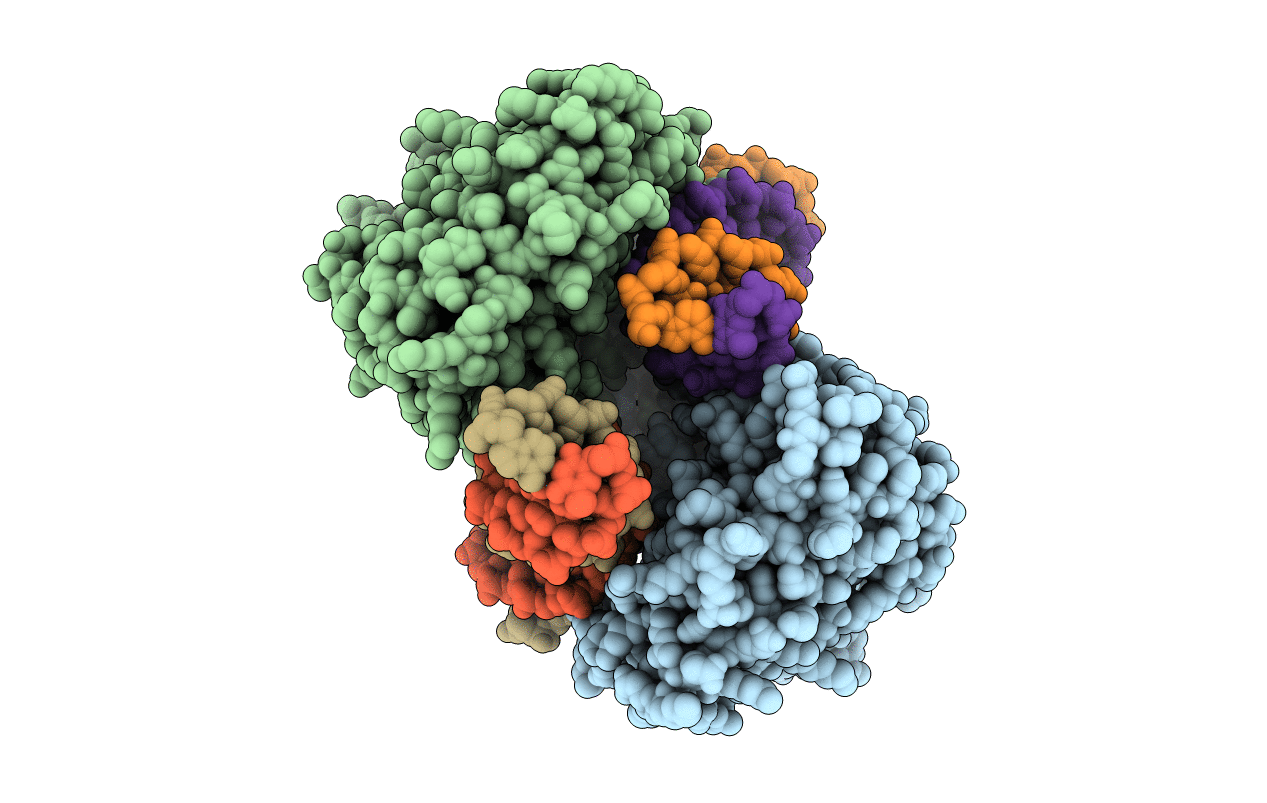
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-12-25 Classification: TRANSFERASE/DNA Ligands: ZN |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2013-12-25 Classification: TRANSFERASE/DNA Ligands: ZN, 1SY |
 |
Solution Structure Of The Core Domain (11-85) Of The Murine Norovirus Vpg Protein
Organism: Murine norovirus 1
Method: SOLUTION NMR Release Date: 2013-03-27 Classification: VIRAL PROTEIN |
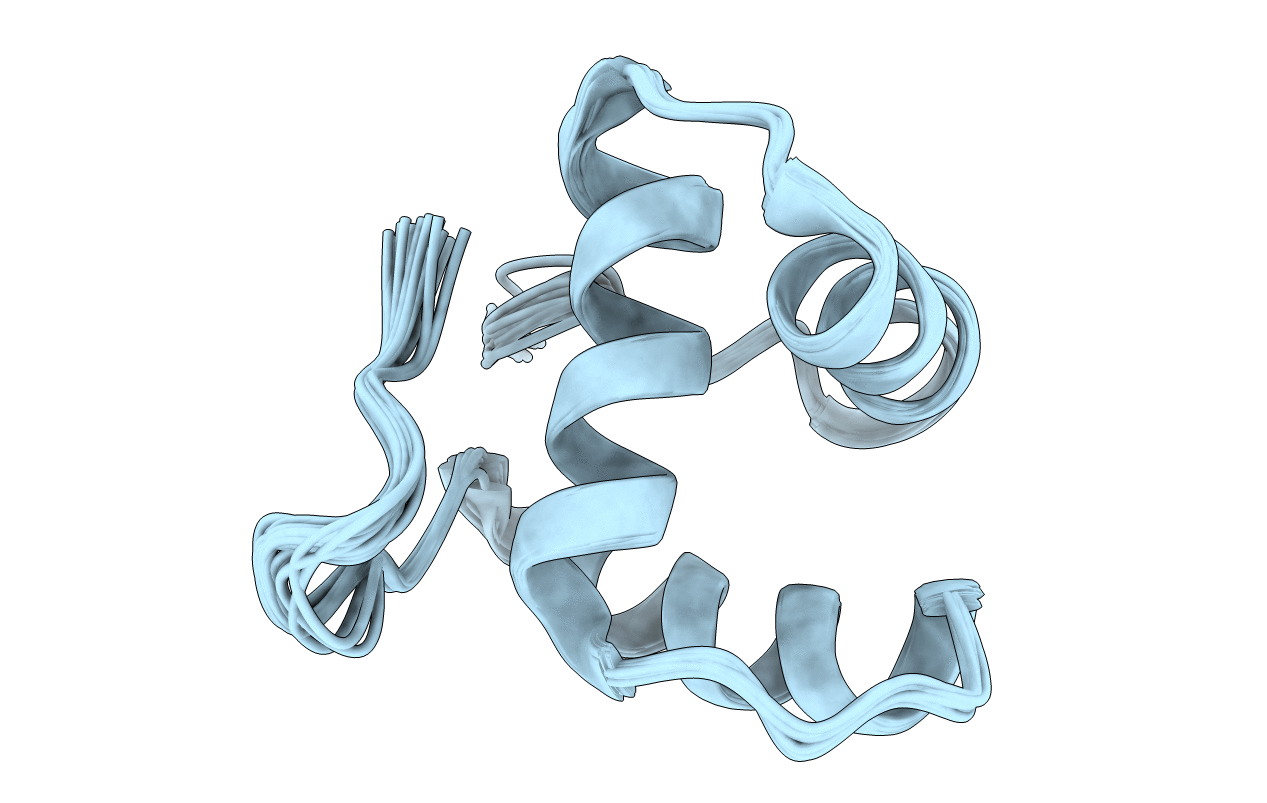 |
Solution Structure Of The Core Domain (10-76) Of The Feline Calicivirus Vpg Protein
Organism: Feline calicivirus
Method: SOLUTION NMR Release Date: 2013-03-27 Classification: VIRAL PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2012-06-13 Classification: MEMBRANE PROTEIN Ligands: C2E, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2012-06-13 Classification: MEMBRANE PROTEIN Ligands: CA |
 |
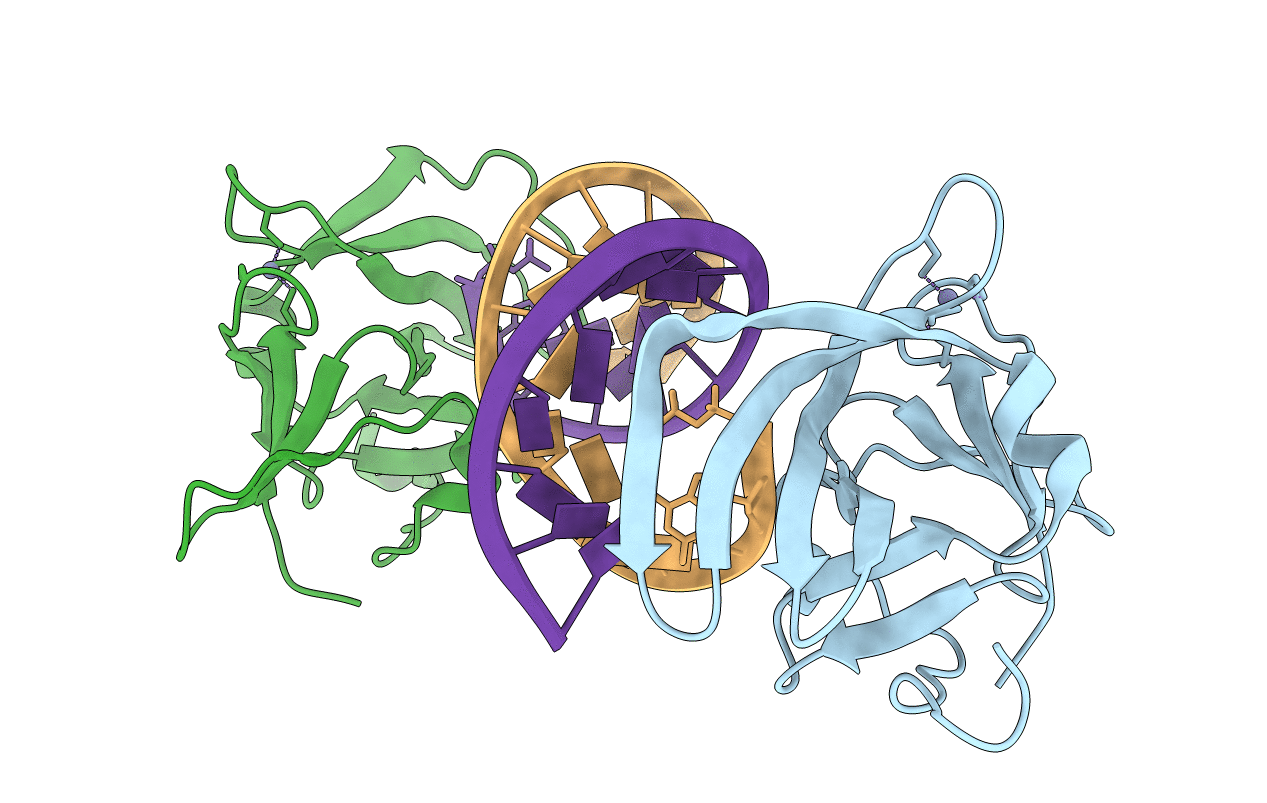
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2010-11-03 Classification: HYDROLASE/RNA Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2010-06-02 Classification: Hydrolase/RNA Ligands: ZN |
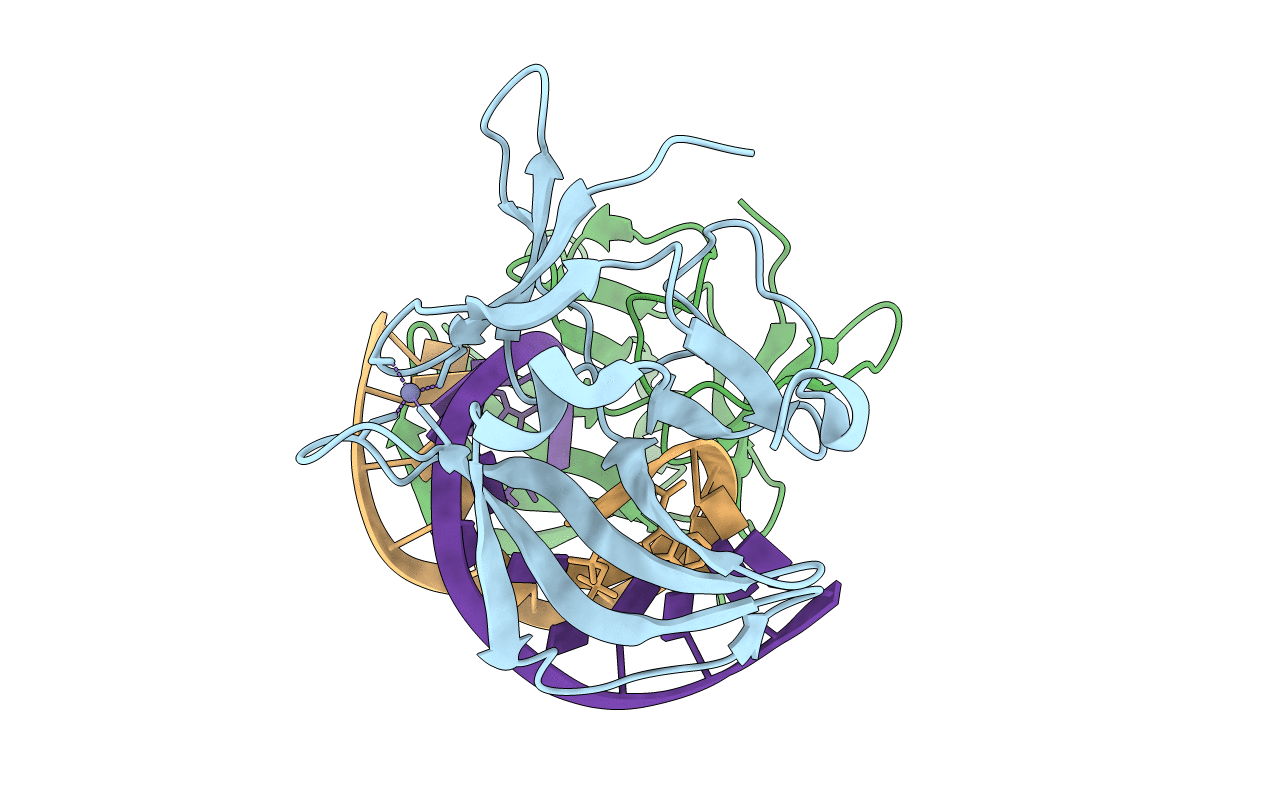 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2010-06-02 Classification: HYDROLASE/RNA Ligands: ZN |
 |
Organism: Sars coronavirus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2007-11-27 Classification: VIRAL PROTEIN |
 |
The Solution Structure Of The Mutant 5'Aug3' Triloop In The Rna Promoter Region Of The Brome Mosaic Virus Genomic (+)-Rna
|
 |
The Solution Structure Of The Stem Loop C 5'Aua3' Triloop Of Brome Mosaic Virus (+) Strand Rna
|

