Search Count: 335
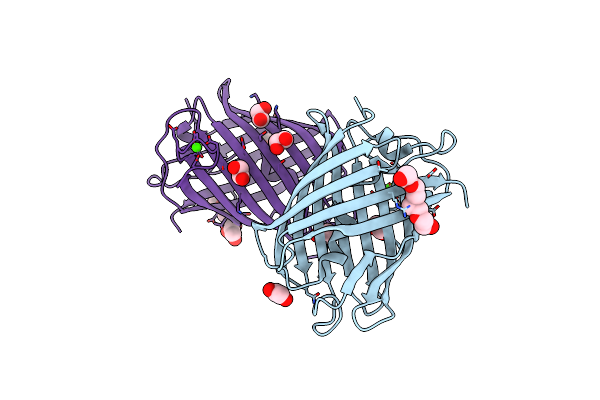 |
Crystal Structure Of Mrfp1 With A Grafted Calcium-Binding Sequence And One Bound Calcium Ion In A Calcium-Containing Solution
Organism: Discosoma, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: FLUORESCENT PROTEIN Ligands: CA, EDO, PG4, PGE |
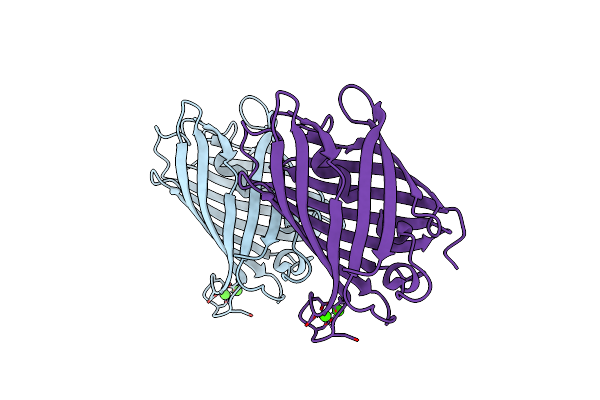 |
Crystal Structure Of Mrfp1 With A Grafted Calcium-Binding Sequence And Two Bound Calcium Ions In A Calcium-Free Solution
Organism: Discosoma, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: FLUORESCENT PROTEIN Ligands: CA |
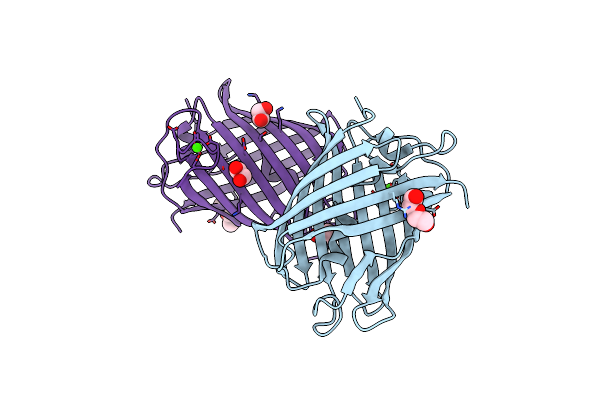 |
Crystal Structure Of Mrfp1 With A Grafted Calcium-Binding Sequence And One Bound Calcium Ion In A Calcium-Free Solution
Organism: Discosoma, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: FLUORESCENT PROTEIN Ligands: CA, PEG, EDO |
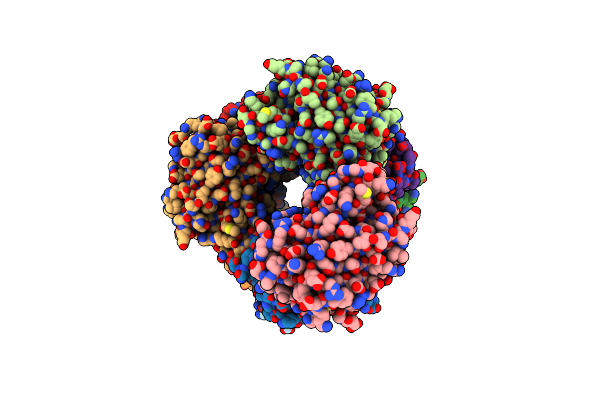 |
Crystal Structure Of The Calcium-Free Mrfp1 With A Grafted Calcium-Binding Sequence
Organism: Discosoma, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: FLUORESCENT PROTEIN |
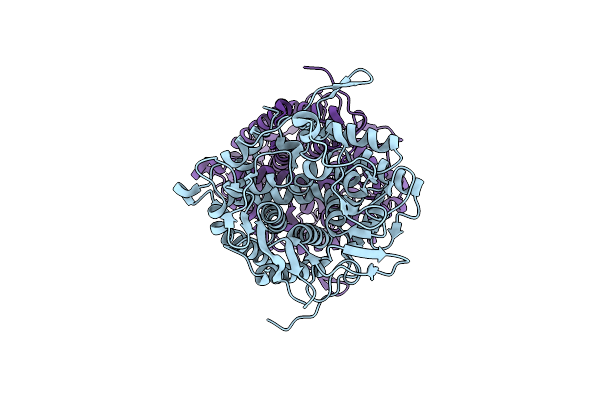 |
Structure Of Beta-1,2-Glucanase From Endozoicomonas Elysicola (Eesgl1, Ligand-Free)
Organism: Endozoicomonas elysicola dsm 22380
Method: X-RAY DIFFRACTION Release Date: 2025-01-22 Classification: HYDROLASE |
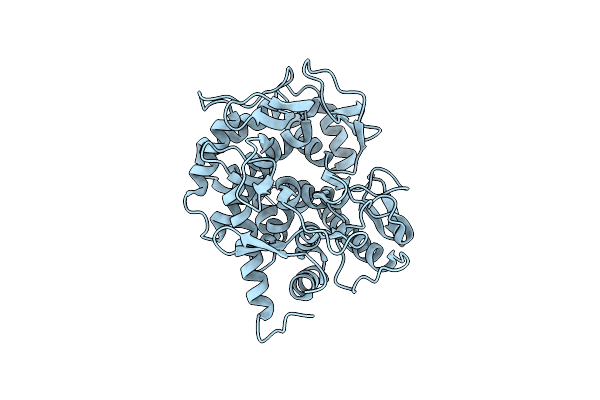 |
Structure Of Beta-1,2-Glucanase From Photobacterium Gaetbulicola (Pgsgl3, Ligand-Free)
Organism: Photobacterium gaetbulicola gung47
Method: X-RAY DIFFRACTION Release Date: 2025-01-22 Classification: HYDROLASE Ligands: CL |
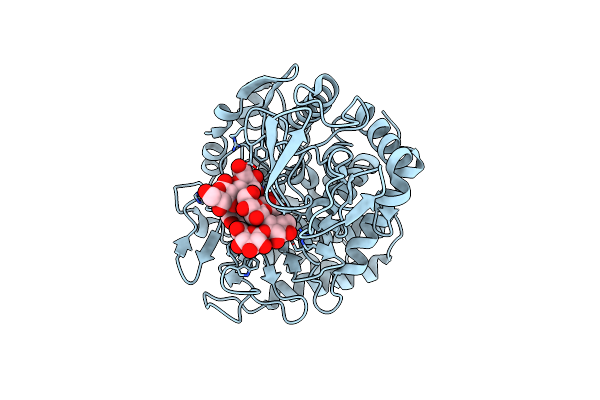 |
Structure Of Beta-1,2-Glucanase From Xanthomonas Campestris Pv. Campestris (Beta-1,2-Glucoheptasaccharide Complex)-E239Q Mutant
Organism: Xanthomonas campestris pv. campestris
Method: X-RAY DIFFRACTION Release Date: 2025-01-22 Classification: HYDROLASE |
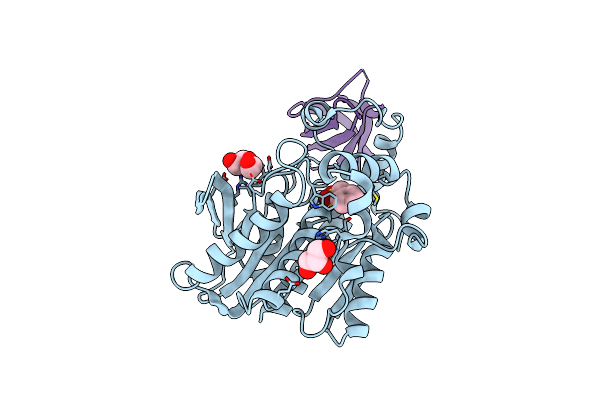 |
Crystal Structure Of The Thermotoga Maritima Cold Shock Protein Mutant Est#13 In Complex With Aacest
Organism: Alicyclobacillus acidocaldarius, Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-15 Classification: PROTEIN BINDING Ligands: PMS, PEG, GOL |
 |
Crystal Structure Of Human Tnf Alpha In Complex With Tnf30(Vhh) Domain Of Ozoralizumab
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2024-09-04 Classification: CYTOKINE |
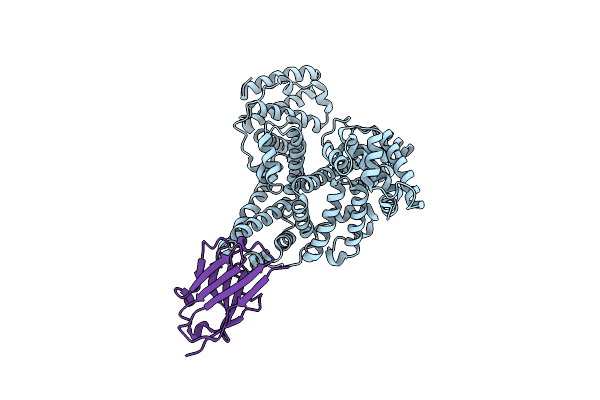 |
Crystal Structure Of Human Serum Albumin In Complex With Alb8(Vhh) Domain Of Ozoralizumab
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-09-04 Classification: TRANSPORT PROTEIN |
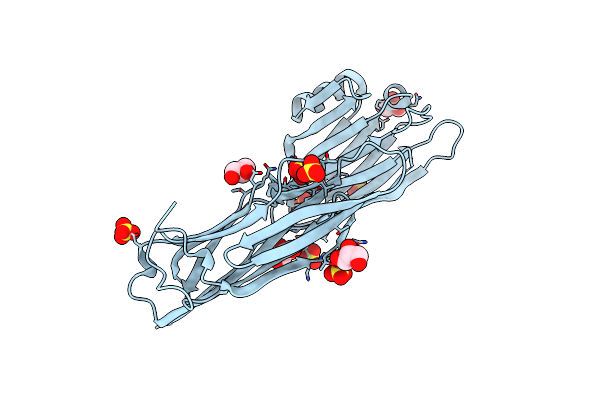 |
Crystal Structure Of The Collagen Binding Domain Of Cnm From Streptococcus Mutans
Organism: Streptococcus mutans
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2024-04-10 Classification: PROTEIN BINDING Ligands: SO4, GOL |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-01-24 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of Sars-Cov-2 Main Protease E166V Mutant In Complex With An Inhibitor Tkb-272
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2023-12-20 Classification: VIRAL PROTEIN Ligands: WOK, PEG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-12-13 Classification: TRANSFERASE Ligands: PE5, N6N, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-12-13 Classification: TRANSFERASE Ligands: MWU |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-12-13 Classification: TRANSFERASE Ligands: PE5, DMS, MWU |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-12-13 Classification: TRANSFERASE Ligands: PE5, MWU, DMS, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-12-13 Classification: TRANSFERASE Ligands: PE5, N7C |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-12-13 Classification: TRANSFERASE Ligands: PE5, N86, DMS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2023-12-13 Classification: TRANSFERASE Ligands: PE5, N8O, DMS |

