Search Count: 17
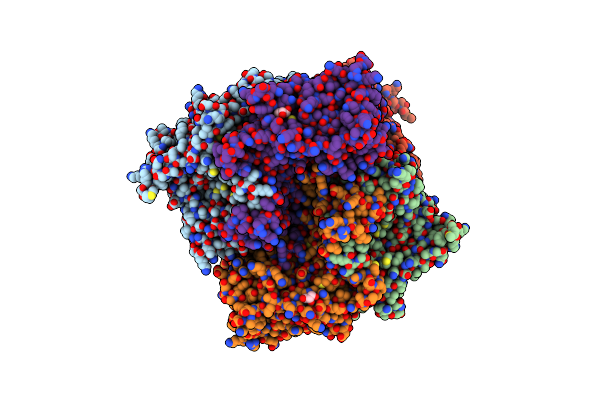 |
Crystal Structure Of Trimethylamine Methyltransferase Mttb From Methanosarcina Barkeri At 2.5 A Resolution
Organism: Methanosarcina barkeri ms
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: GOL, NA |
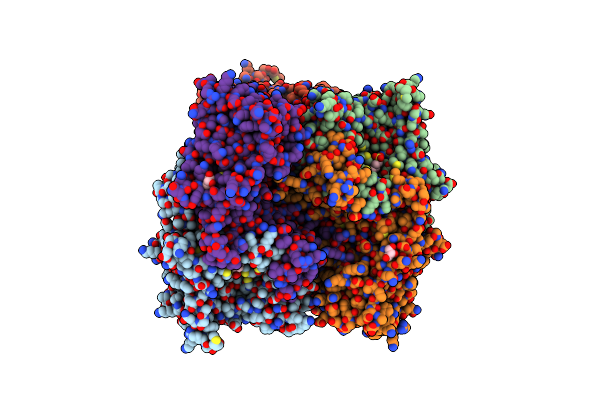 |
Organism: Methanosarcina barkeri ms
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: BG3, NA, GOL |
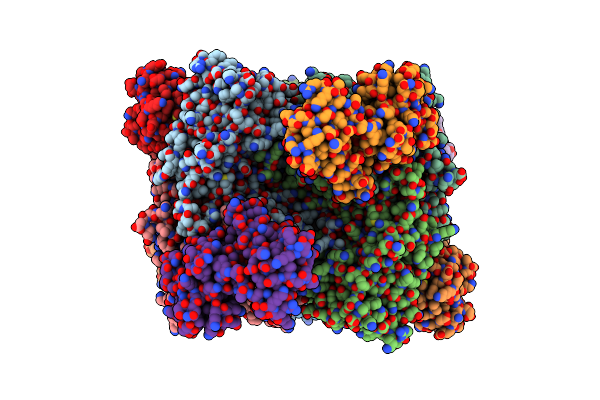 |
Organism: Methanosarcina barkeri ms
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: GOL, HCB |
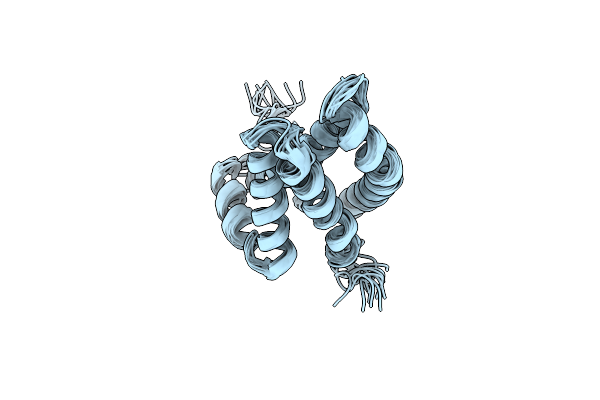 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2020-03-04 Classification: MEMBRANE PROTEIN |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2019-03-13 Classification: TRANSFERASE Ligands: PG4, NA |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2019-03-13 Classification: TRANSFERASE Ligands: ATP, NA, RIB |
 |
Structure Of Arabidopsis Thaliana Ribokinase Complexed With Atp And Magnesium Ion
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-03-13 Classification: TRANSFERASE Ligands: ATP, MG, NA |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2019-02-06 Classification: HYDROLASE Ligands: 1PE, PEG |
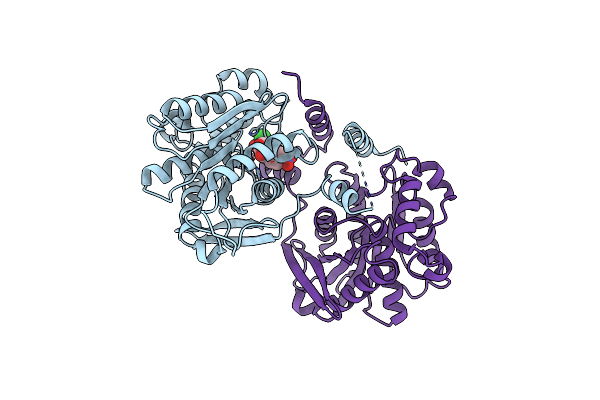 |
Crystal Structure Of Chloramphenicol-Metabolizaing Enzyme Estdl136-Chloramphenicol Complex
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-02-06 Classification: HYDROLASE Ligands: CLM |
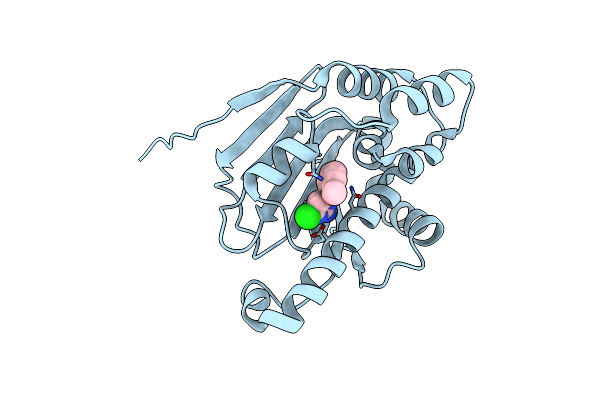 |
Optimization Of Potent, Selective, And Orally Bioavailable Pyrrolodinopyrimidine-Containing Inhibitors Of Heat Shock Protein 90. Identification Of Development Candidate 2-Amino-4-{4-Chloro-2-[2-(4-Fluoro-1H-Pyrazol-1-Yl)Ethoxy]-6-Methylphenyl}-N-(2,2-Difluoropropyl)-5,7-Dihydro-6H-Pyrrolo[3,4-D]Pyrimidine-6-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-04-27 Classification: Chaperone/Chaperone inhibitor Ligands: WOE |
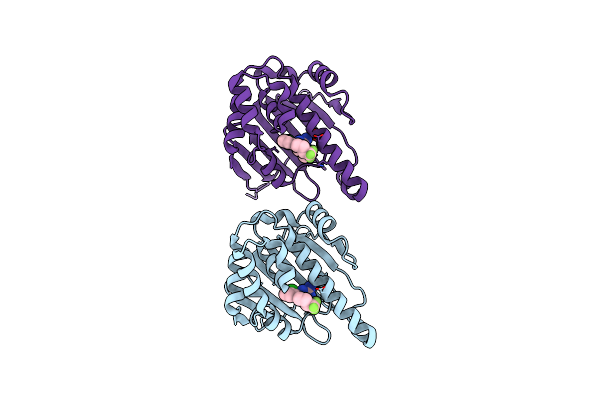 |
Optimization Of Potent, Selective, And Orally Bioavailable Pyrrolodinopyrimidine-Containing Inhibitors Of Heat Shock Protein 90. Identification Of Development Candidate 2-Amino-4-{4-Chloro-2-[2-(4-Fluoro-1H-Pyrazol-1-Yl)Ethoxy]-6-Methylphenyl}-N-(2,2-Difluoropropyl)-5,7-Dihydro-6H-Pyrrolo[3,4-D]Pyrimidine-6-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-04-27 Classification: Chaperone/Chaperone inhibitor Ligands: FU5 |
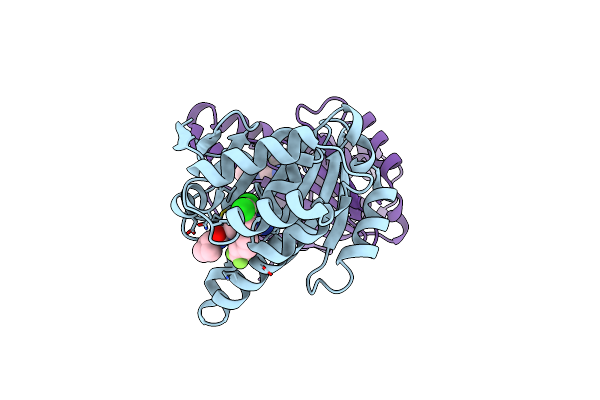 |
Optimization Of Potent, Selective, And Orally Bioavailable Pyrrolodinopyrimidine-Containing Inhibitors Of Heat Shock Protein 90. Identification Of Development Candidate 2-Amino-4-{4-Chloro-2-[2-(4-Fluoro-1H-Pyrazol-1-Yl)Ethoxy]-6-Methylphenyl}-N-(2,2-Difluoropropyl)-5,7-Dihydro-6H-Pyrrolo[3,4-D]Pyrimidine-6-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2011-04-27 Classification: Chaperone/Chaperone inhibitor Ligands: FU3 |
 |
Optimization Of Potent, Selective, And Orally Bioavailable Pyrrolodinopyrimidine-Containing Inhibitors Of Heat Shock Protein 90. Identification Of Development Candidate 2-Amino-4-{4-Chloro-2-[2-(4-Fluoro-1H-Pyrazol-1-Yl)Ethoxy]-6-Methylphenyl}-N-(2,2-Difluoropropyl)-5,7-Dihydro-6H-Pyrrolo[3,4-D]Pyrimidine-6-Carboxamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-04-27 Classification: Chaperone/Chaperone inhibitor Ligands: FU7, PO4 |
 |
Hsp90 N-Terminal Domain In Complex With 1-{4-[(2R)-1-(5-Chloro-2,4-Dihydroxybenzoyl)Pyrrolidin-2-Yl]Benzyl}-3,3-Difluoropyrrolidinium
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2009-09-29 Classification: CHAPERONE Ligands: BD0, PO4 |
 |
Organism: Methanosarcina barkeri
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-10-19 Classification: TRANSFERASE Ligands: BG5 |
 |
Organism: Methanosarcina barkeri
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-10-19 Classification: TRANSFERASE Ligands: BG4 |
 |
Organism: Methanosarcina barkeri
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-10-19 Classification: TRANSFERASE Ligands: SO4, BG3 |

