Search Count: 239
 |
Photoactivation In Bacteriophytochromes, Reference (Dark) Structure For The 3 Ps Time Point
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: 3Q8, BEN |
 |
Photoactivation In Bacteriophytochrome, High Resolution Cryo Structure In The Dark.
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, P33 |
 |
Photoactivation In Bacteriophytochromes, Reference (Dark) Structure For The 100 Ps Time Point
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: EL5, BEN |
 |
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: BLA, BEN |
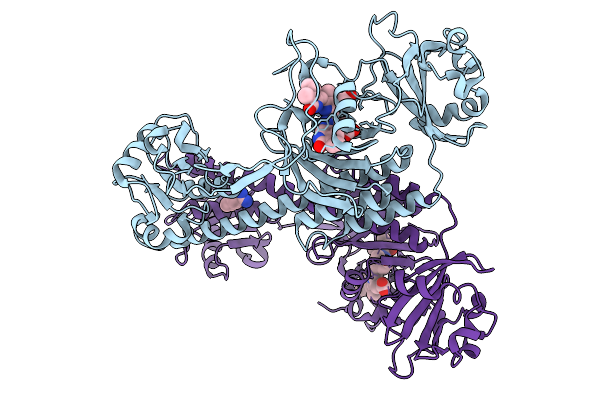 |
Organism: Stigmatella aurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: SIGNALING PROTEIN Ligands: BLA, BEN |
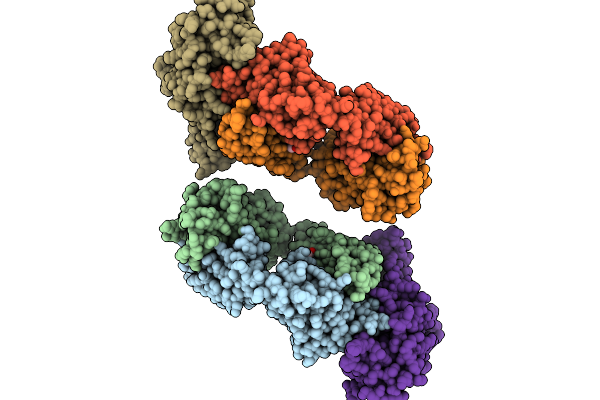 |
Crystal Structure Of Sars-Cov-2 Consp Rbd In Complex With Neutralizing Antibody Cc25.4 Fab
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: IMMUNE SYSTEM Ligands: EDO |
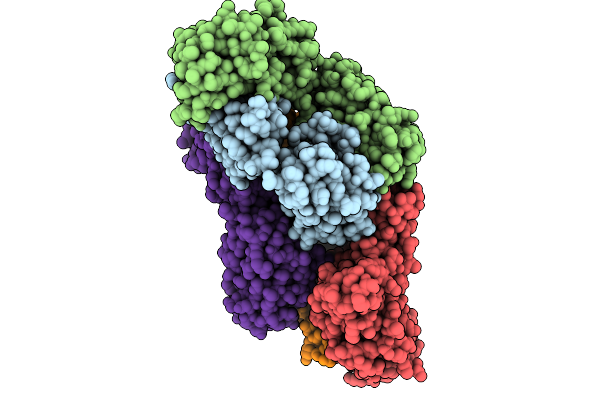 |
Crystal Structure Of Sars-Cov-2 Consp Rbd In Complex With Antibodies Pdi222 Fab And Cova1-16 Fab
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: IMMUNE SYSTEM |
 |
Organism: Pseudomonas aeruginosa
Method: SOLUTION NMR Release Date: 2025-06-18 Classification: VIRAL PROTEIN |
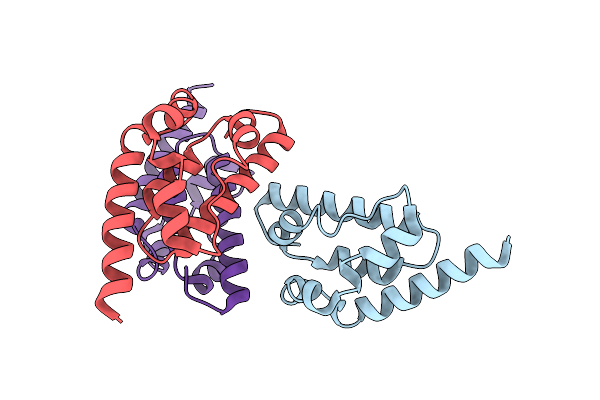 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: VIRAL PROTEIN |
 |
Cryo-Em Structure Of Colibactin Assembly Line Polyketide Synthase Clbc (Apo State)
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: BIOSYNTHETIC PROTEIN |
 |
Cryo-Em Structure Of Colibactin Assembly Line Polyketide Synthase Clbc (Acp-Bound State)
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: TRANSFERASE |
 |
Cryo-Em Structure Of Colibactin Assembly Line Polyketide Synthase Clbi (Apo State)
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: TRANSFERASE |
 |
Cryo-Em Structure Of Colibactin Assembly Line Polyketide Synthase Clbi Ks-At Didomain Crosslinked With Clbi Acp
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: TRANSFERASE |
 |
Cryo-Em Structure Of Colibactin Assembly Line Polyketide Synthase Clbi Ks-At Didomain Crosslinked With Its Precursor Module, Clbh
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: TRANSFERASE |
 |
Organism: Lama glama
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: IMMUNE SYSTEM Ligands: GOL |
 |
Organism: Homo sapiens, Lama glama
Method: X-RAY DIFFRACTION Release Date: 2025-05-21 Classification: IMMUNE SYSTEM Ligands: NAG, SCN, GOL, SO4 |
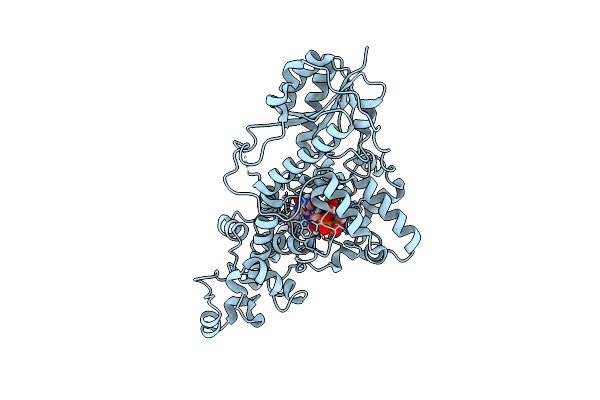 |
Organism: Chlamydomonas reinhardtii cc3269
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2025-05-14 Classification: FLAVOPROTEIN Ligands: FAD, CL |
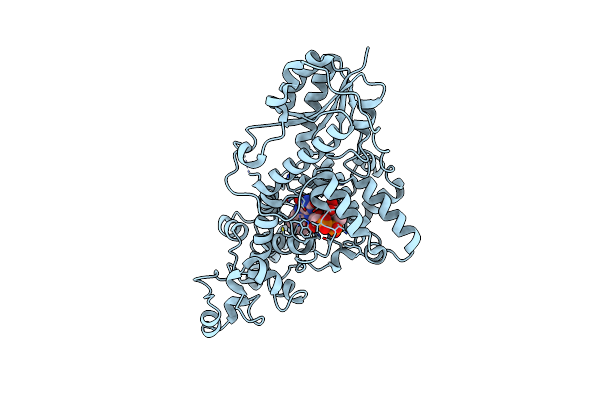 |
Sfx Structure Of Cracry 10 Ns After Photoexcitation Of The Oxidized Protein
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-05-14 Classification: FLAVOPROTEIN Ligands: FAD, CL |
 |
Sfx Structure Of Cracry 30 Ns After Photoexcitation Of The Oxidized Protein
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-05-14 Classification: FLAVOPROTEIN Ligands: FAD, CL |
 |
Sfx Structure Of Cracry 100 Ns After Photoexcitation Of The Oxidized Protein
Organism: Chlamydomonas reinhardtii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-05-14 Classification: FLAVOPROTEIN Ligands: FAD, CL |

