Search Count: 36
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-02-03 Classification: TRANSFERASE |
 |
Crystal Structure Of Human Quinolinate Phosphoribosyltransferase In Complex With The Reactant Quinolinate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.09 Å Release Date: 2016-02-03 Classification: TRANSFERASE Ligands: NTM |
 |
Crystal Structure Of Human Quinolinate Phosphoribosyltransferase In Complex With The Product Nicotinate Mononucleotide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-02-03 Classification: TRANSFERASE Ligands: NCN |
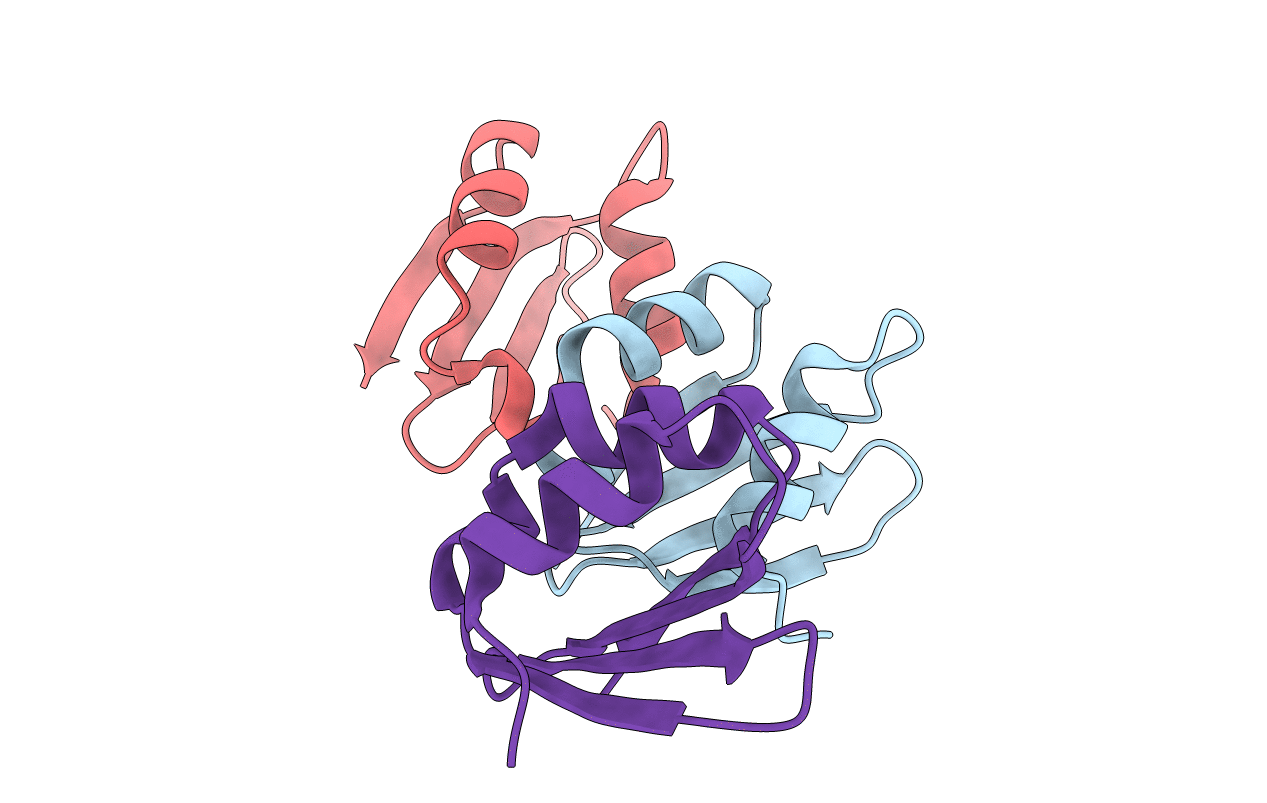 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2015-04-01 Classification: HYDROLASE |
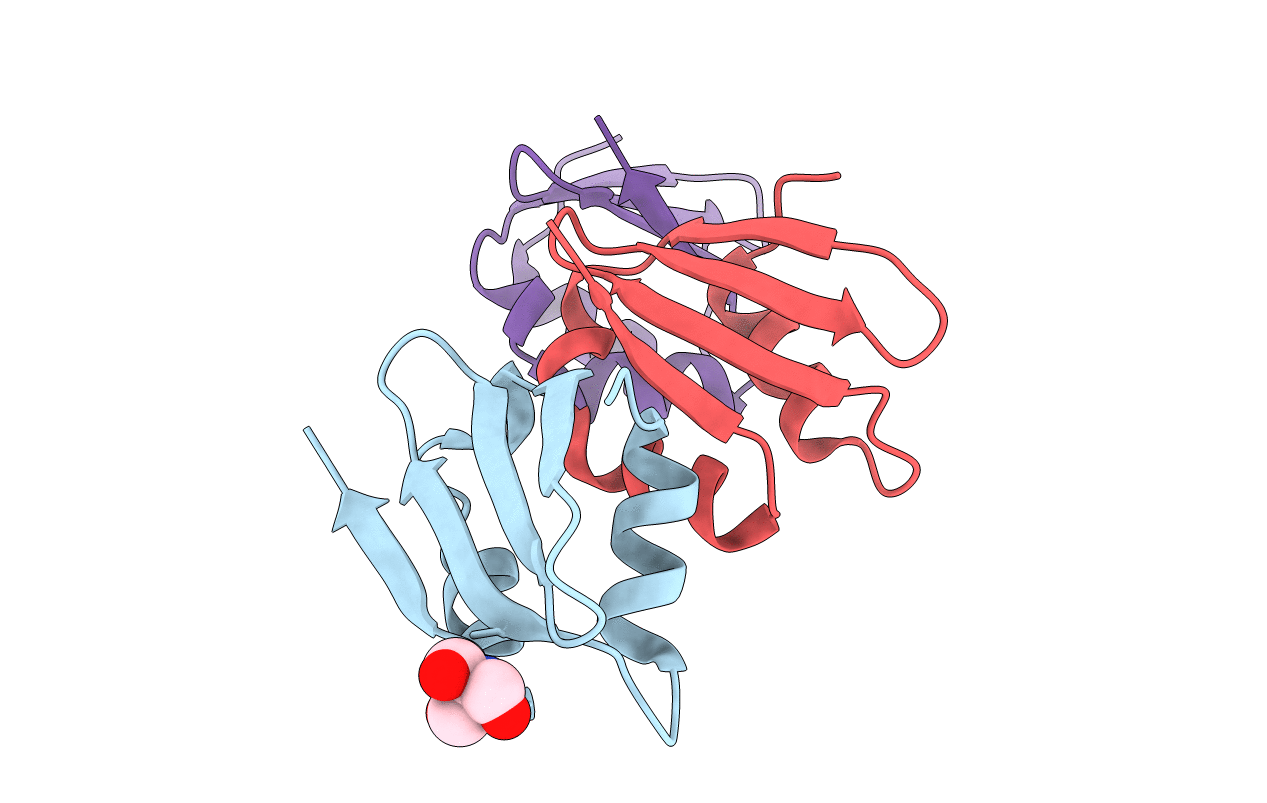 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2014-09-17 Classification: HYDROLASE Ligands: TRS |
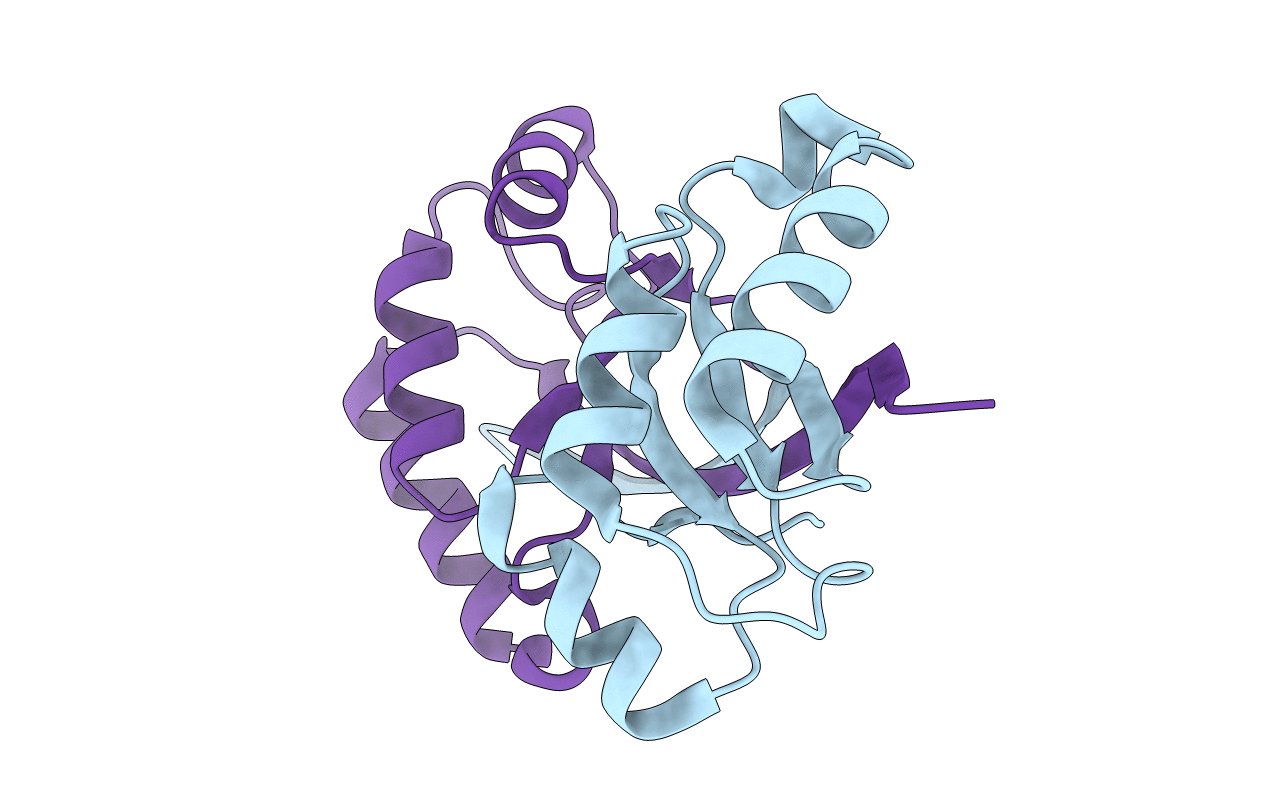 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2013-10-23 Classification: PROTEIN BINDING |
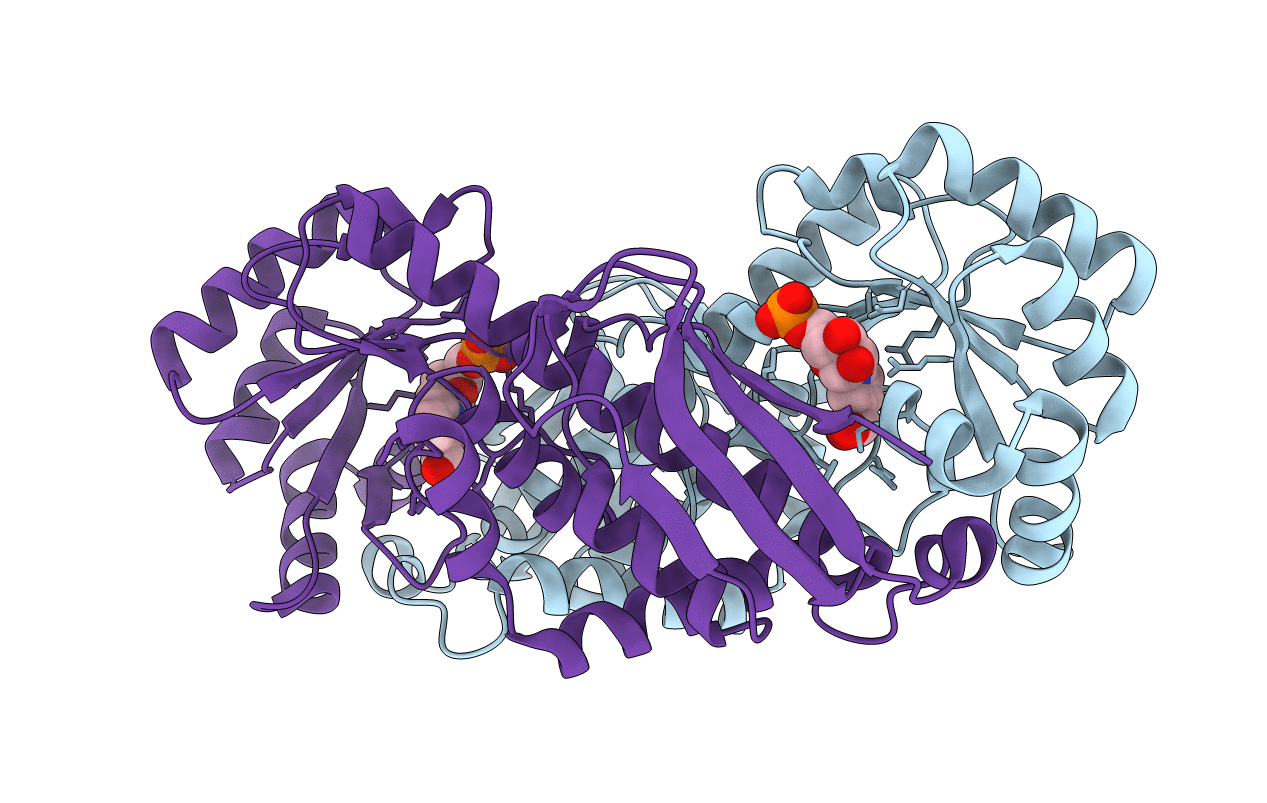 |
Crystal Structure Of Sus Scrofa Quinolinate Phosphoribosyltransferase In Complex With Nicotinate Mononucleotide
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-05-01 Classification: TRANSFERASE Ligands: NCN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-09-08 Classification: LIGASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-09-08 Classification: LIGASE/SIGNALING PROTEIN |
 |
Crystal Structure Of Helicobacter Pylori Mine, A Cell Division Topological Specificity Factor
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-05-05 Classification: CELL CYCLE |
 |
Crystal Structure Of Helicobacter Pylori Mine, A Cell Division Topological Specificity Factor
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2010-05-05 Classification: CELL CYCLE |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-02-16 Classification: SIGNALING PROTEIN/PROTEIN BINDING |
 |
Crystal Structure Of Rattus Norvegicus Visfatin/Pbef/Nampt In Complex With An Fk866-Based Inhibitor
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2009-08-18 Classification: TRANSFERASE Ligands: IS1 |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2008-09-09 Classification: HYDROLASE |
 |
Crystal Structure Of Helicobacter Pylori Atp Dependent Protease, Ftsh Adp Complex
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2008-09-09 Classification: HYDROLASE Ligands: ADP |
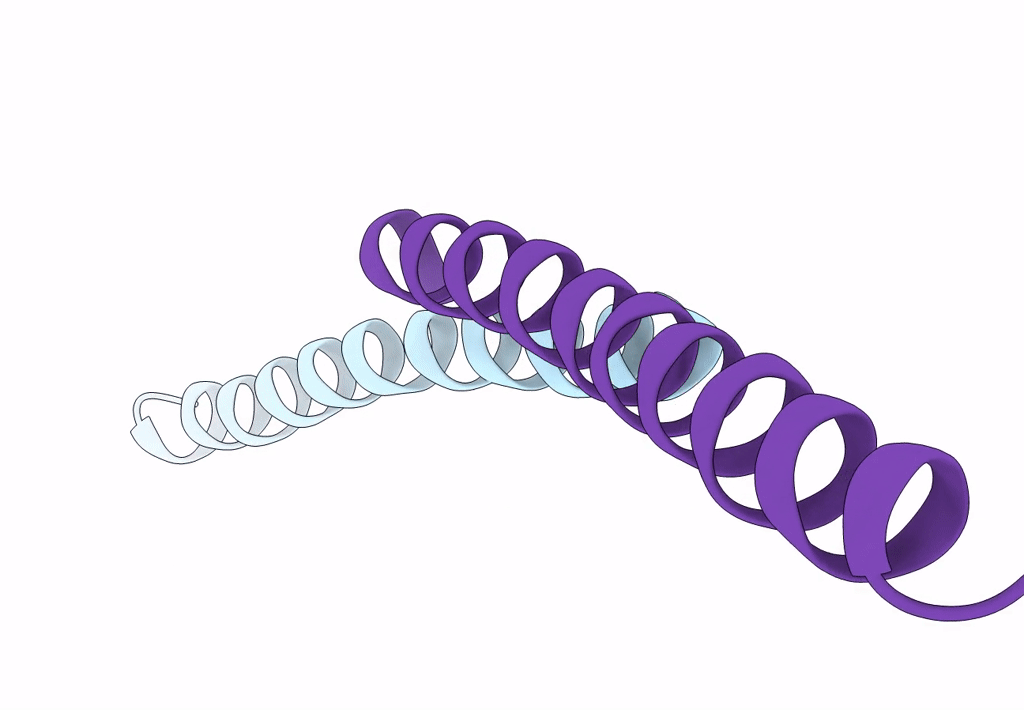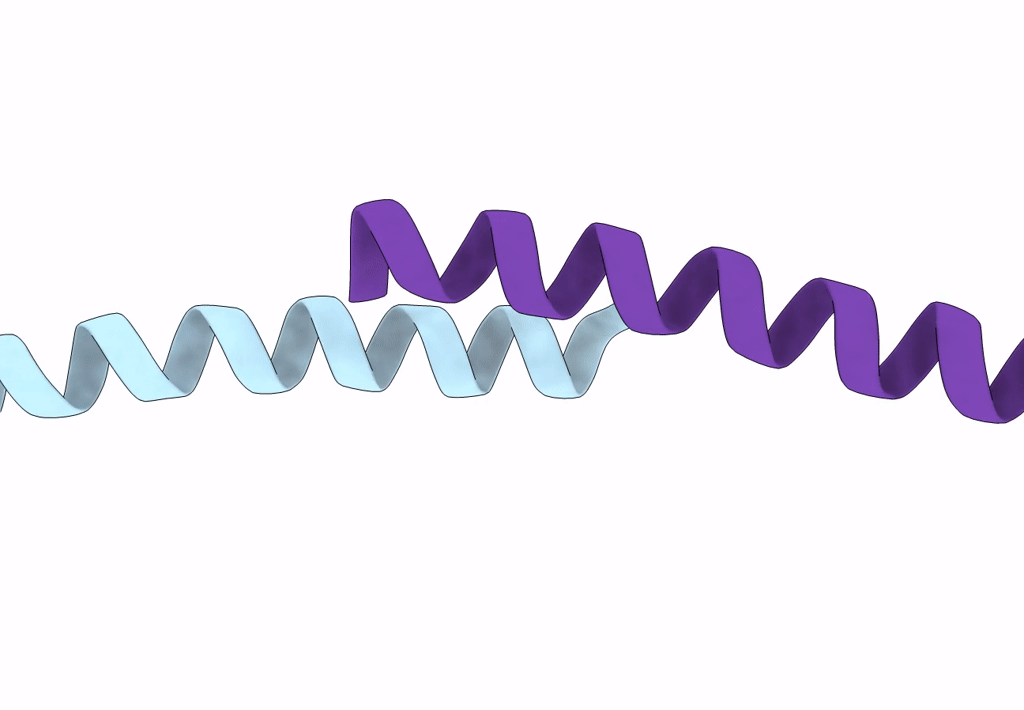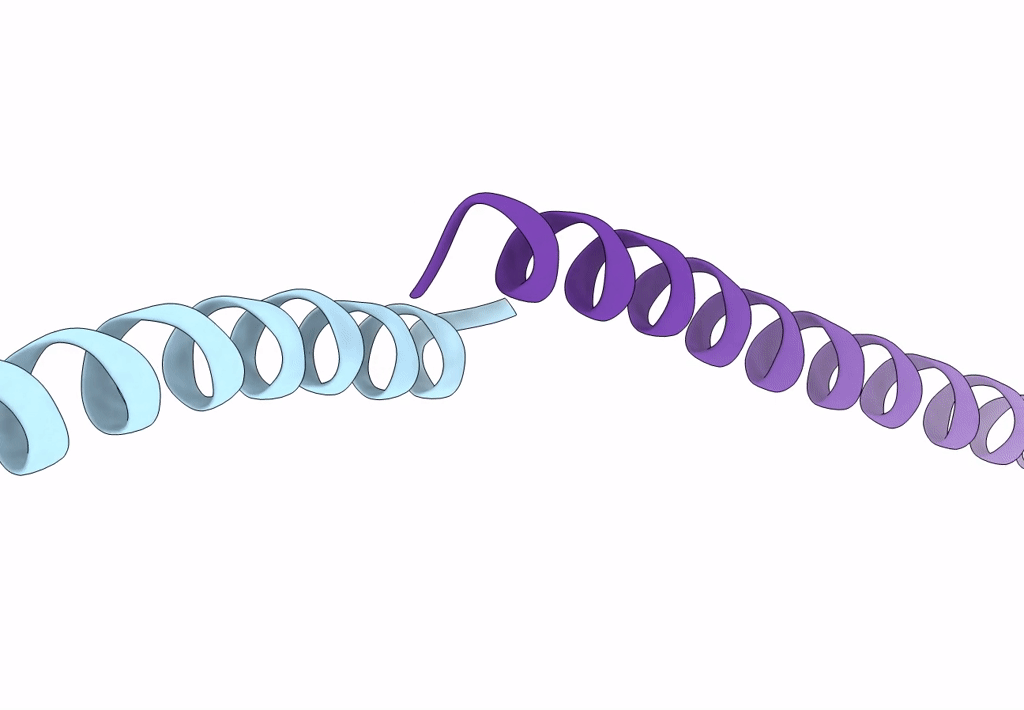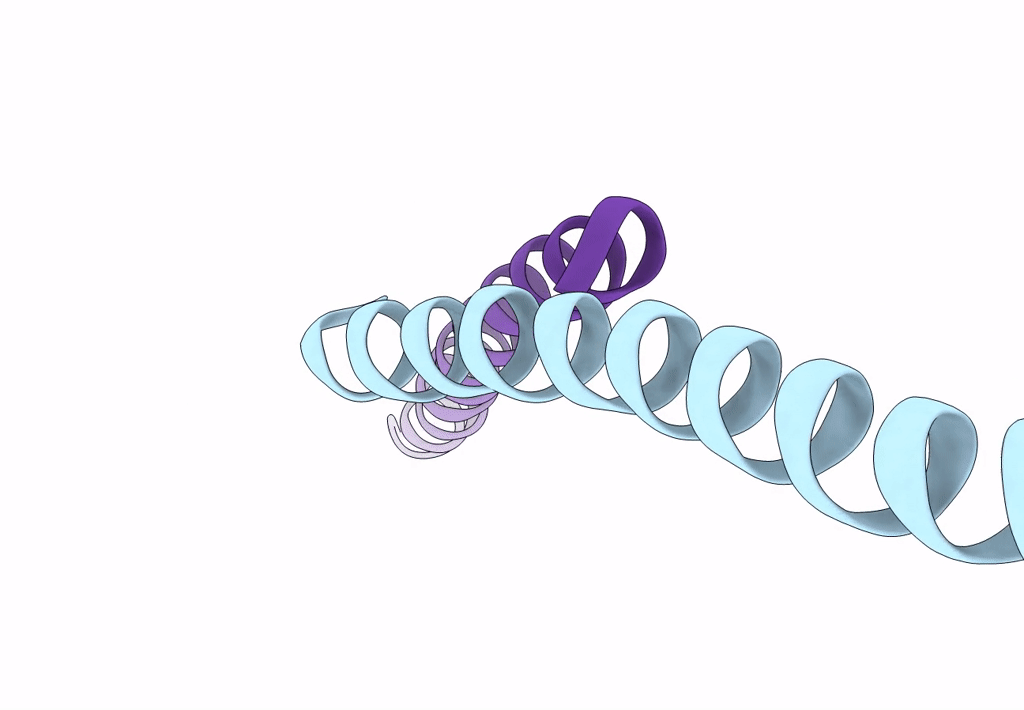 |
Crystal Structure Of The Leucine Zipper Domain Of Small-Conductance Ca2+-Activated K+ (Skca) Channel From Rattus Norvegicus
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-04-29 Classification: MEMBRANE PROTEIN |
 |
Crystal Structure Of Bacillus Subtilis Codw, A Non-Canonical Hslv-Like Peptidase With An Impaired Catalytic Apparatus
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2008-03-25 Classification: HYDROLASE |
 |
Crystal Structure Of Bacillus Subtilis Codw, A Non-Canonical Hslv-Like Peptidase With An Impaired Catalytic Apparatus
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-03-25 Classification: HYDROLASE |
 |
Crystal Structure Of The Glur0 Ligand-Binding Core From Nostoc Punctiforme In Complex With (L)-Glutamate
Organism: Nostoc punctiforme
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-01-22 Classification: TRANSPORT PROTEIN Ligands: GLU |
 |
Crystal Structure Of Udp-N-Acetylenolpyruvylglucosamine Reductase (Murb) From Thermus Caldophilus
Organism: Thermus caldophilus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2006-12-19 Classification: OXIDOREDUCTASE Ligands: CA, FAD |

