Search Count: 84
 |
Organism: Escherichia coli, Homo sapiens, Macaca mulatta, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-05-29 Classification: SIGNALING PROTEIN |
 |
Full Agonist- And Positive Allosteric Modulator-Bound Mu-Type Opioid Receptor-G Protein Complex
Organism: Escherichia coli, Homo sapiens, Macaca mulatta, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-05-29 Classification: SIGNALING PROTEIN Ligands: VV9 |
 |
Crystal Structure Of Glycoside Hydrolase Family 11 Beta-Xylanase From Streptomyces Olivaceoviridis E-86
Organism: Streptomyces olivaceoviridis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-12-30 Classification: HYDROLASE Ligands: CL |
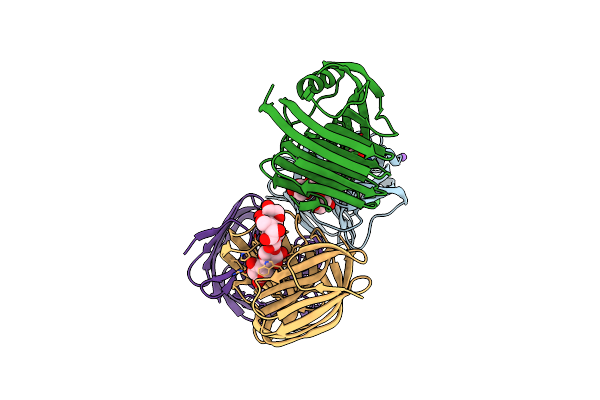 |
Crystal Structure Of Glycoside Hydrolase Family 11 Beta-Xylanase From Streptomyces Olivaceoviridis E-86 In Complex With Alpha-L-Arabinofuranosyl Xylotetraose
Organism: Streptomyces olivaceoviridis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-12-30 Classification: HYDROLASE Ligands: CL, NA |
 |
Crystal Structure Of Glycoside Hydrolase Family 11 Beta-Xylanase From Streptomyces Olivaceoviridis E-86 In Complex With 4-O-Methyl-Alpha-D-Glucuronopyranosyl Xylotetraose
Organism: Streptomyces olivaceoviridis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-12-30 Classification: HYDROLASE Ligands: CL, NA |
 |
Crystal Structure Of Exo-Beta-1,3-Galactanase From Phanerochaete Chrysosporium Pc1,3Gal43A Apo Form
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-11-04 Classification: HYDROLASE Ligands: NAG, CA, CIT |
 |
Crystal Structure Of Exo-Beta-1,3-Galactanase From Phanerochaete Chrysosporium Pc1,3Gal43A With Galactose
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-11-04 Classification: HYDROLASE Ligands: NAG, CA, GLA, GAL, ACY, ACT, GOL |
 |
Crystal Structure Of Exo-Beta-1,3-Galactanase From Phanerochaete Chrysosporium Pc1,3Gal43A E208Q With Beta-1,3-Galactotriose
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-11-04 Classification: HYDROLASE Ligands: NAG, CA, GAL |
 |
Crystal Structure Of Exo-Beta-1,3-Galactanase From Phanerochaete Chrysosporium Pc1,3Gal43A E208A With Beta-1,3-Galactotriose
Organism: Phanerochaete chrysosporium
Method: X-RAY DIFFRACTION Release Date: 2020-11-04 Classification: HYDROLASE Ligands: CA, ACT, GOL, NAG |
 |
Organism: Cellulomonas fimi (strain atcc 484 / dsm 20113 / jcm 1341 / nbrc 15513 / ncimb 8980 / nctc 7547)
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-08-26 Classification: HYDROLASE |
 |
Crystal Structure Of Human Estrogen-Related Receptor Gamma Ligand Binding Domain Complex With Bpa-Monof
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2020-05-13 Classification: TRANSCRIPTION Ligands: CW6 |
 |
Crystal Structure Of Covalent Glycosyl-Enzyme Intermediate Of Xylanase Mutant (T82A, N127S, And E128H) From Streptomyces Olivaceoviridis E-86
Organism: Streptomyces olivaceoviridis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-08-09 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of Michaelis Complex Of Xylanase Mutant (T82A, N127S, And E128H) From Streptomyces Olivaceoviridis E-86
Organism: Streptomyces olivaceoviridis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-08-09 Classification: HYDROLASE |
 |
Crystal Structure Of Coxyl43, Gh43 Beta-Xylosidase/Alpha-Arabinofuranosidase From A Compost Microbial Metagenome, Calcium-Free Form.
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-03-15 Classification: HYDROLASE |
 |
Crystal Structure Of Coxyl43, Gh43 Beta-Xylosidase/Alpha-Arabinofuranosidase From A Compostmicrobial Metagenome, Calcium-Bound Form
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-03-15 Classification: HYDROLASE Ligands: CA, NA, GOL, ACT |
 |
Crystal Structure Of Coxyl43, Gh43 Beta-Xylosidase/Alpha-Arabinofuranosidase From A Compost Microbial Metagenome In Complex With Xylotriose, Calcium-Free Form.
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-03-15 Classification: HYDROLASE |
 |
Crystal Structure Of Coxyl43, Gh43 Beta-Xylosidase/Alpha-Arabinofuranosidase From A Compostmicrobial Metagenome In Complex With Xylotriose, Calcium-Bound Form
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-03-15 Classification: HYDROLASE Ligands: CA, NA, XYS, XYP, ACT |
 |
Crystal Structure Of Coxyl43, Gh43 Beta-Xylosidase/Alpha-Arabinofuranosidase From A Compostmicrobial Metagenome In Complex With L-Arabinose, Calcium-Free Form
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-03-15 Classification: HYDROLASE |
 |
Crystal Structure Of Coxyl43, Gh43 Beta-Xylosidase/Alpha-Arabinofuranosidase From A Compostmicrobial Metagenome In Complex With L-Arabinose, Calcium-Bound Form
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-03-15 Classification: HYDROLASE Ligands: CA, NA, ARA, ACT |
 |
Crystal Structure Of Coxyl43, Gh43 Beta-Xylosidase/Alpha-Arabinofuranosidase From A Compostmicrobial Metagenome In Complex With L-Arabinose And Xylotriose, Calcium-Free Form
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-03-15 Classification: HYDROLASE |

