Search Count: 39
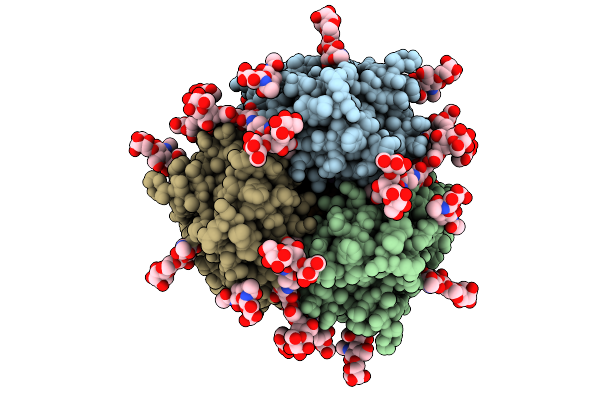 |
Cryo-Em Structure Of Croatia 2023 H3N2 Influenza A Hemagglutinin (Ha) Trimer
Organism: Influenza a virus
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG |
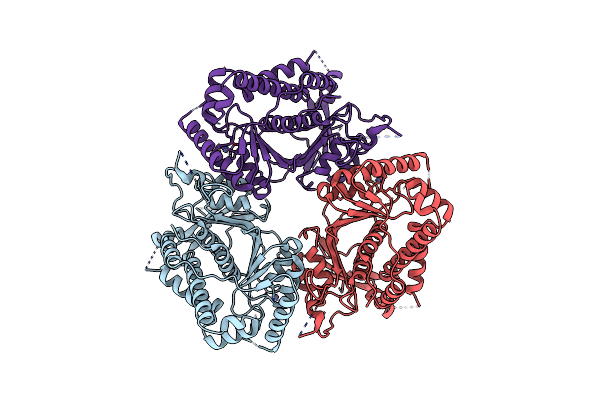 |
Cryo-Em Structure Of Human Cdadc1 Inactive Mutant (E400A): Trimer Without A Ligand
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: HYDROLASE Ligands: ZN |
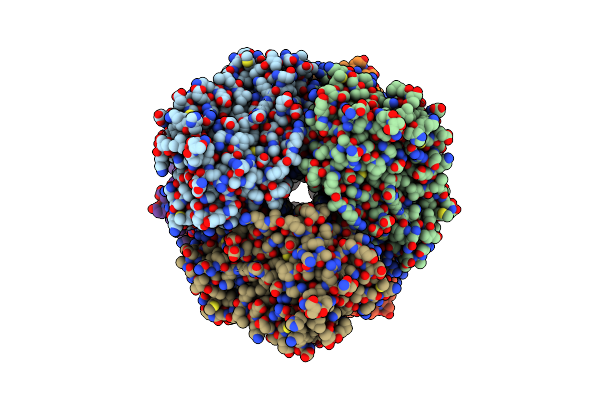 |
Cryo-Em Structure Of Human Cdadc1 Inactive Mutant (E400A): Hexamer Without A Ligand
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: HYDROLASE Ligands: ZN |
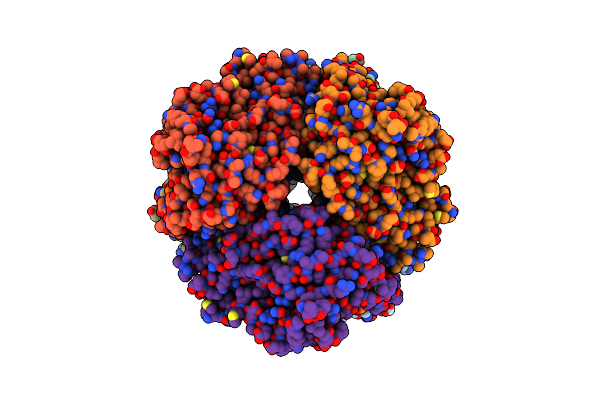 |
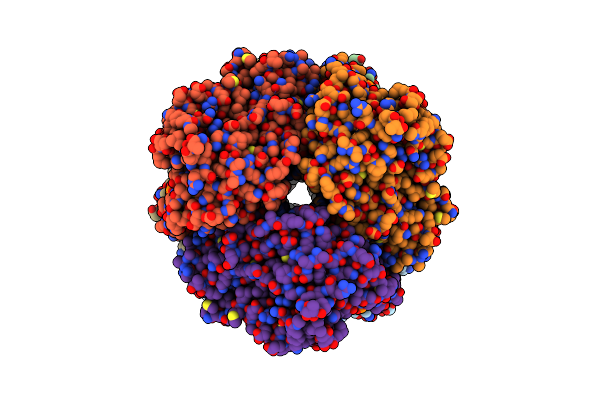
Cryo-Em Structure Of Human Cdadc1 Inactive Mutant (E400A): Hexamer With Dctp Bound In The Active Site
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: HYDROLASE Ligands: ZN, DCP |
 |
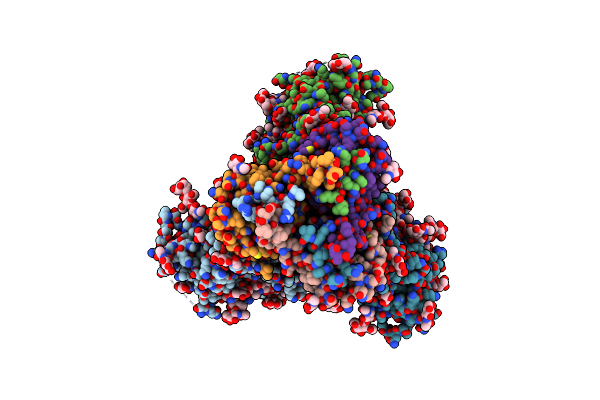
Cryo-Em Structure Of Human Cdadc1 Inactive Mutant (E400A): Hexamer With 5Mdctp Bound In The Active Site
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: HYDROLASE Ligands: ZN, A1I2I |
 |
Cryo-Em Structure Of Pgdm1400 Fab Bound To Hiv-1 Bg505 Ds-Sosip.664 Env Trimer
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: VIRAL PROTEIN Ligands: NAG |
 |
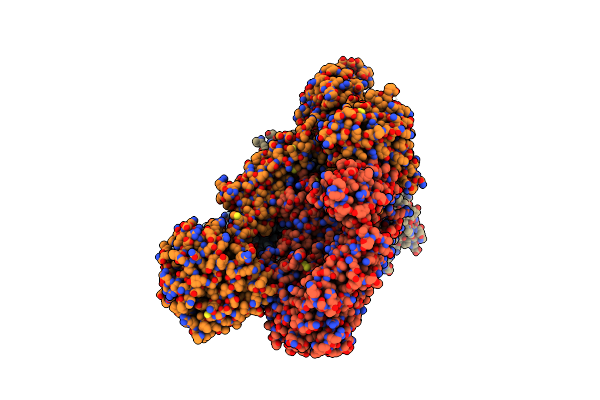
Cryo-Em Structure Of Pgt145 R100As Fab Bound To Hiv-1 Bg505 Ds-Sosip.664 Env Trimer
Organism: Human immunodeficiency virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: VIRAL PROTEIN Ligands: NAG, PO4 |
 |
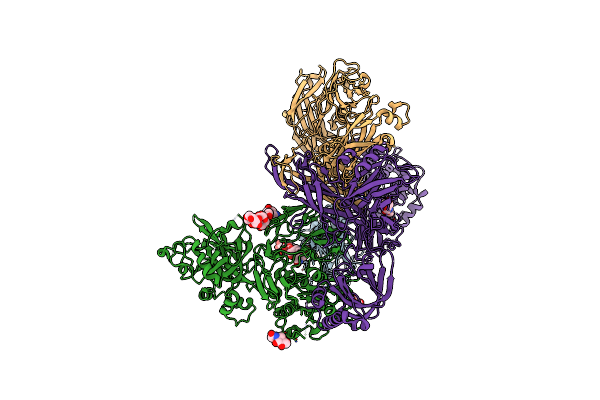
Organism: Lutzomyia longipalpis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: IMMUNE SYSTEM Ligands: MG, NAG |
 |
Organism: Lutzomyia longipalpis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: IMMUNE SYSTEM Ligands: NAG |
 |
Organism: Lutzomyia longipalpis
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2023-08-09 Classification: BLOOD CLOTTING Ligands: GOL, NAG, BR |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-12-14 Classification: RNA Ligands: ZN, SO4, GOL, EDO, ACT |
 |
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2022-06-01 Classification: RIBOSOME/INHIBITOR Ligands: MG, MQ6, ZN |
 |
Organism: Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2022-06-01 Classification: RIBOSOME/INHIBITOR Ligands: MG, MQ6, ZN |
 |
Crystal Structure Of A Soluble Variant Of Full-Length Human Apobec3G (Ph 7.6)
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-12-23 Classification: HYDROLASE/DNA Ligands: ZN, CL, GOL |
 |
Crystal Structure Of A Soluble Variant Of Full-Length Human Apobec3G (Ph 8.0)
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2020-12-23 Classification: HYDROLASE/DNA Ligands: ZN |
 |
Crystal Structure Of A Soluble Variant Of Full-Length Human Apobec3G (Ph 9.0)
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2020-12-23 Classification: HYDROLASE/DNA Ligands: ZN |
 |
Crystal Structure Of Apobec3G Catalytic Domain Complex With Substrate Ssdna
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2018-07-18 Classification: HYDROLASE/DNA Ligands: ZN, GOL |
 |
Organism: Thermococcus kodakarensis (strain atcc baa-918 / jcm 12380 / kod1)
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2018-06-20 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Organism: Thermococcus kodakarensis (strain atcc baa-918 / jcm 12380 / kod1)
Method: X-RAY DIFFRACTION Resolution:3.24 Å Release Date: 2018-06-20 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Organism: Thermococcus chitonophagus
Method: X-RAY DIFFRACTION Resolution:3.72 Å Release Date: 2018-05-02 Classification: HYDROLASE Ligands: CO, SO4 |

