Search Count: 16
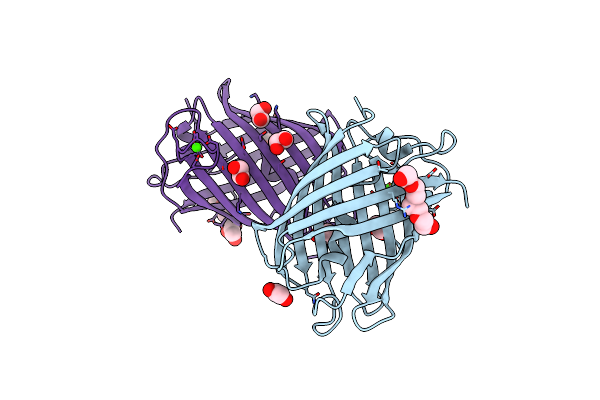 |
Crystal Structure Of Mrfp1 With A Grafted Calcium-Binding Sequence And One Bound Calcium Ion In A Calcium-Containing Solution
Organism: Discosoma, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: FLUORESCENT PROTEIN Ligands: CA, EDO, PG4, PGE |
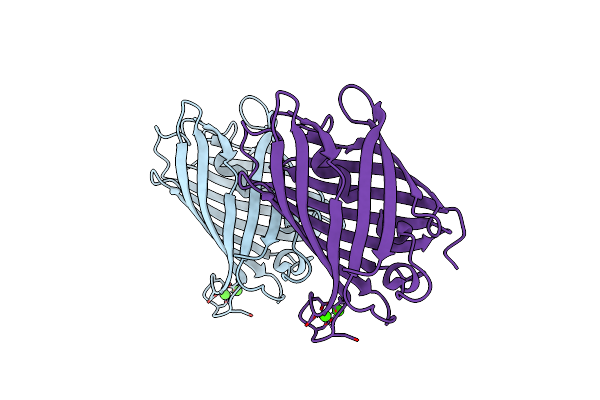 |
Crystal Structure Of Mrfp1 With A Grafted Calcium-Binding Sequence And Two Bound Calcium Ions In A Calcium-Free Solution
Organism: Discosoma, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: FLUORESCENT PROTEIN Ligands: CA |
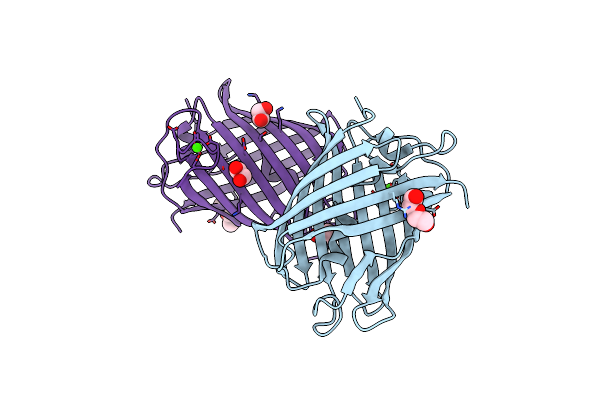 |
Crystal Structure Of Mrfp1 With A Grafted Calcium-Binding Sequence And One Bound Calcium Ion In A Calcium-Free Solution
Organism: Discosoma, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: FLUORESCENT PROTEIN Ligands: CA, PEG, EDO |
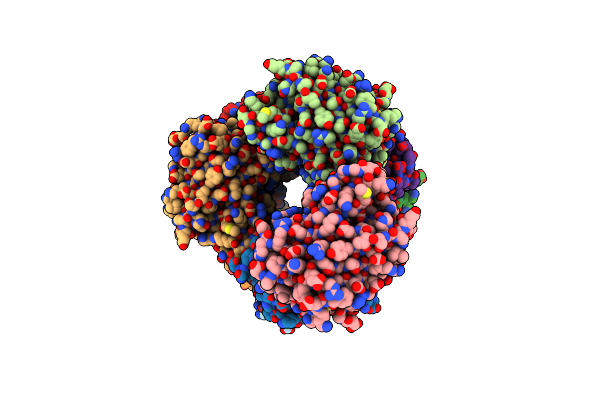 |
Crystal Structure Of The Calcium-Free Mrfp1 With A Grafted Calcium-Binding Sequence
Organism: Discosoma, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-04 Classification: FLUORESCENT PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-11-25 Classification: RNA/NUCLEIC ACID |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-11-25 Classification: RNA/NUCLEIC ACID Ligands: CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-04-06 Classification: PROTEIN TRANSPORT Ligands: GOL, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-04-06 Classification: PROTEIN TRANSPORT Ligands: CA, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2016-04-06 Classification: PROTEIN TRANSPORT Ligands: CA, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2016-04-06 Classification: PROTEIN TRANSPORT Ligands: CA, CL |
 |
Solution Structure Of The A' Domain Of Thermophilic Fungal Protein Disulfide (Oxidized Form, 303K)
|
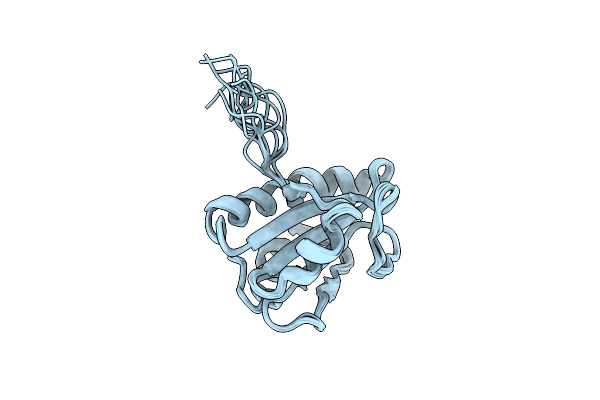 |
Solution Structure Of The A' Domain Of Thermophilic Fungal Protein Disulfide (Reduced Form, 303K)
|
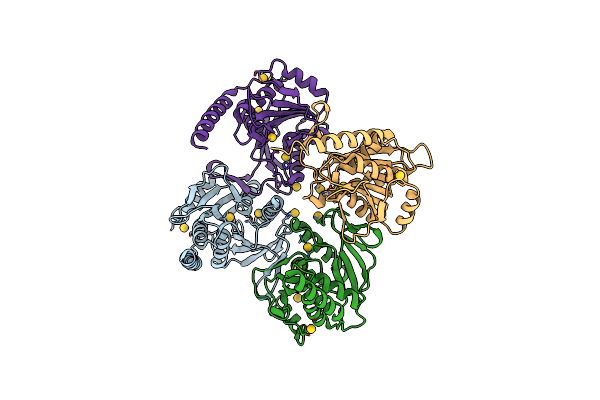 |
Crystal Structure Of Pyrococcus Furiosus Pbab, An Archaeal Proteasome Activator
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-04-03 Classification: PROTEIN BINDING Ligands: AU |
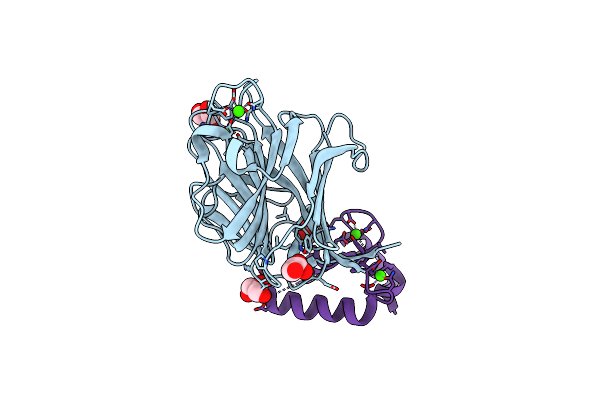 |
Crystal Structure Of Mcfd2 In Complex With Carbohydrate Recognition Domain Of Ergic-53
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2010-01-05 Classification: PROTEIN TRANSPORT Ligands: CA, GOL |
 |
Solution Structure Of The A' Domain Of Thermophilic Fungal Protein Disulfide Isomerase
|
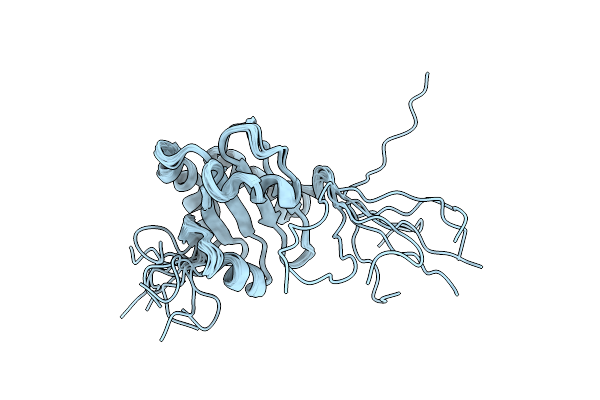 |
Solution Structure Of The B' Domain Of Thermophilic Fungal Protein Disulfide Isomerase
|

