Search Count: 192
 |
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: METAL BINDING PROTEIN Ligands: NI, NAD, 1BO |
 |
X-Ray Structure Of Clostridioides Difficile Autolysin Acd33800 Catalytic Domain
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Release Date: 2025-10-15 Classification: ANTIMICROBIAL PROTEIN |
 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE |
 |
X-Ray Structure Of Enterobacter Cloaca Transaldolase In Complex With D-Fructose-6-Phosphate.
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: F6R |
 |
Organism: Thermus thermophilus (strain atcc 27634 / dsm 579 / hb8)
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: TRANSFERASE Ligands: SO4, EDO |
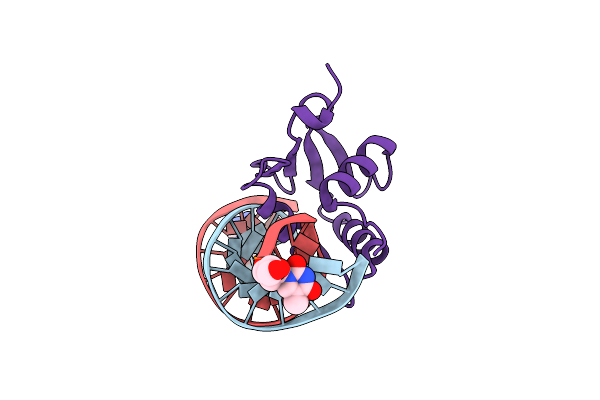 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: DNA BINDING PROTEIN Ligands: 3D1, THM |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: DNA BINDING PROTEIN Ligands: 3D1, THM, PG4 |
 |
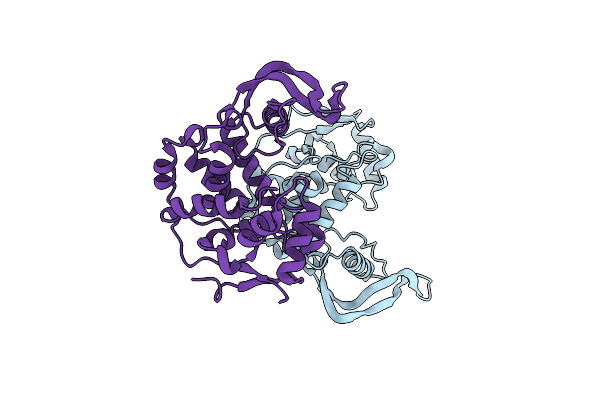
X-Ray Structure Of Clostridioides Difficile Endolysin Ecd09610 Glucosaminidase Domain.
Organism: Clostridioides difficile (strain 630)
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2024-05-15 Classification: HYDROLASE |
 |
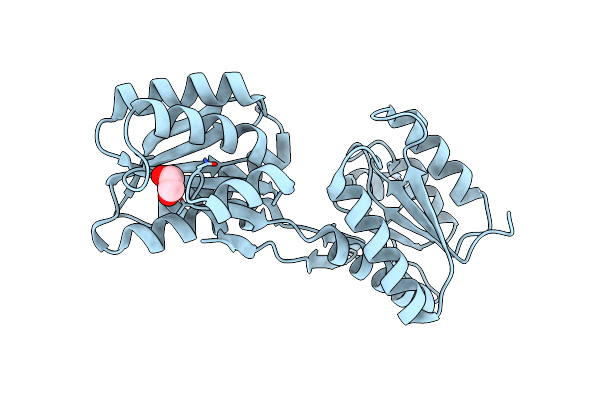
X-Ray Structure Of Clostridium Perfringens Autolysin Catalytic Domain In The P1 Form
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-05-15 Classification: HYDROLASE |
 |
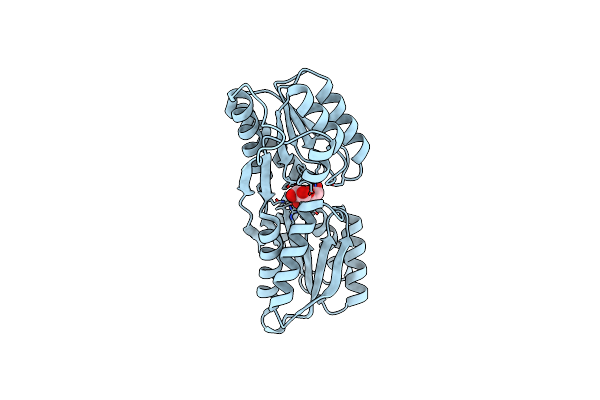
X-Ray Structure Of Enterobacter Cloacae Allose-Binding Protein In Free Form
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2023-10-25 Classification: SUGAR BINDING PROTEIN Ligands: EDO |
 |
X-Ray Structure Of Enterobacter Cloacae Allose-Binding Protein In Complex With D-Allose
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2023-10-25 Classification: SUGAR BINDING PROTEIN Ligands: ALL |
 |
X-Ray Structure Of Enterobacter Cloacae Allose-Binding Protein In Complex With D-Ribose
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2023-10-25 Classification: SUGAR BINDING PROTEIN Ligands: RIP |
 |
X-Ray Structure Of Enterobacter Cloacae Allose-Binding Protein In Complex With D-Psicose
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2023-10-25 Classification: SUGAR BINDING PROTEIN Ligands: WEB |
 |
X-Ray Structure Of Clostridium Perfringens Pili Protein B Collagen-Binding Domains
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2023-06-07 Classification: PROTEIN FIBRIL Ligands: ACT |
 |
X-Ray Structure Of Clostridium Perfringens Pili Protein B N-Terminal Domain
Organism: Clostridium perfringens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-06-07 Classification: PROTEIN FIBRIL |
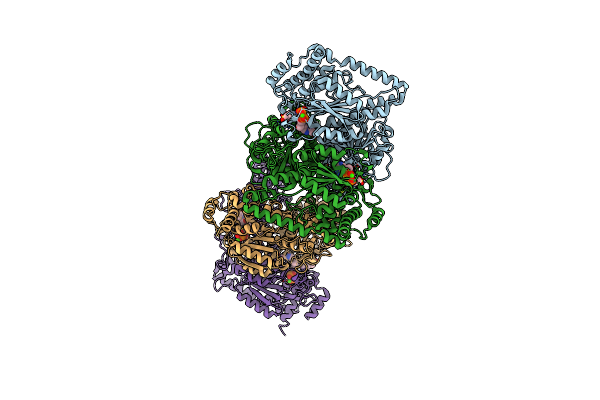 |
X-Ray Structure Of Thermus Thermophilus Hb8 Transketorase In Complex With Tpp And Mes
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2022-12-07 Classification: TRANSFERASE Ligands: TPP, CA, MES |
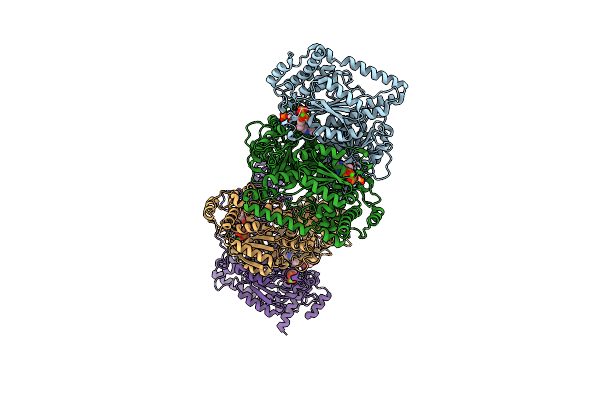 |
X-Ray Structure Ofthermus Thermophilus Hb8 Transketorase Demonstrate In Complex With Tpp And D-Erythrose-4-Phosphate
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-12-07 Classification: TRANSFERASE Ligands: TPP, CA, E4P |
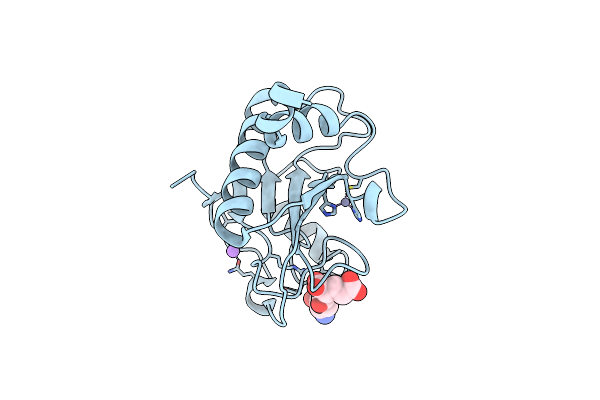 |
Organism: Clostridium perfringens (strain 13 / type a)
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-05-04 Classification: HYDROLASE Ligands: GLU, ZN, NA |
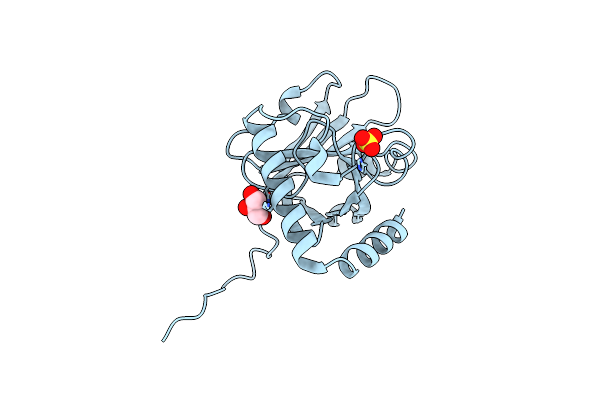 |
X-Ray Structure Of The Intermolecular Complex Of Clostridium Perfringens Sortase C With The C-Terminal Cell Wall Sorting Signal Motif.
Organism: Clostridium perfringens (strain sm101 / type a)
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2021-06-02 Classification: TRANSFERASE Ligands: SO4, GOL |
 |
X-Ray Structure Of Clostridium Perfringens Sortase C With The C-Terminal Cell Wall Sorting Motif.
Organism: Clostridium perfringens (strain sm101 / type a)
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2021-06-02 Classification: TRANSFERASE |

