Search Count: 64
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2017-12-20 Classification: HYDROLASE |
 |
Crystal Structures Of Human Orexin 2 Receptor Bound To The Selective Antagonist Empa Determined By Serial Femtosecond Crystallography At Sacla
Organism: Homo sapiens, Pyrococcus abyssi (strain ge5 / orsay)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-12-13 Classification: SIGNALING PROTEIN Ligands: 7MA, OLA, 1PE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-12-06 Classification: HYDROLASE |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2017-12-06 Classification: HYDROLASE |
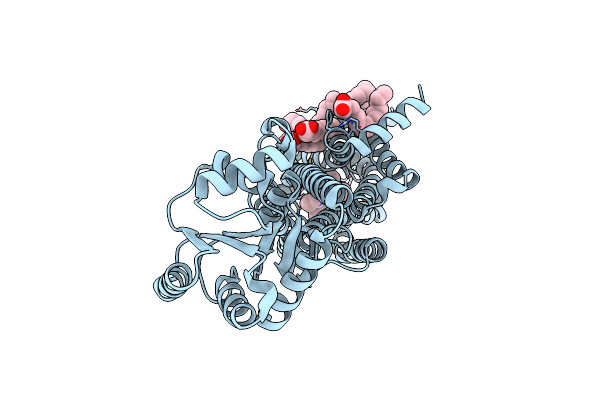 |
Crystal Structure Of Human Orexin 2 Receptor Bound To The Selective Antagonist Empa Determined By The Synchrotron Light Source At Spring-8.
Organism: Homo sapiens, Pyrococcus abyssi ge5
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2017-11-29 Classification: SIGNALING PROTEIN Ligands: 7MA, OLA, 1PE |
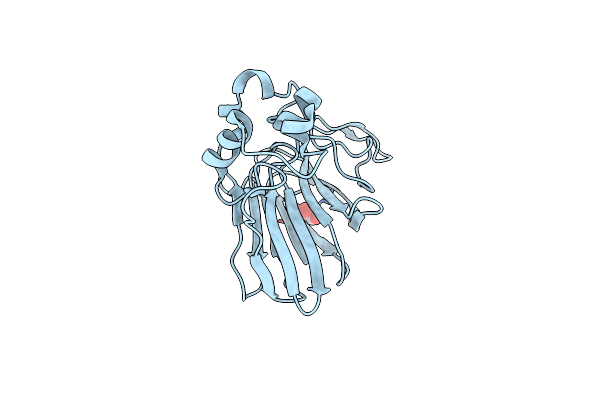 |
Organism: Thaumatococcus daniellii
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2017-11-29 Classification: PLANT PROTEIN Ligands: TLA |
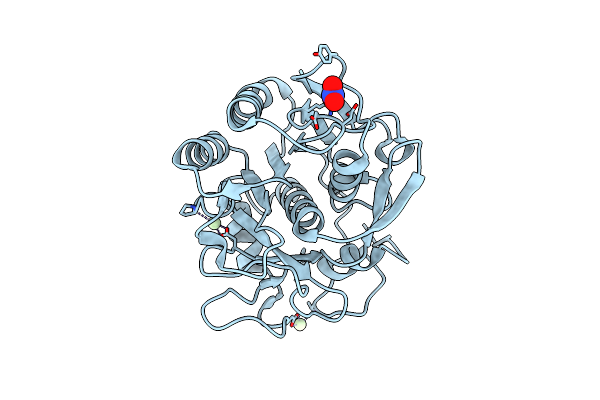 |
Organism: Engyodontium album
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-11-29 Classification: HYDROLASE Ligands: NO3, PR |
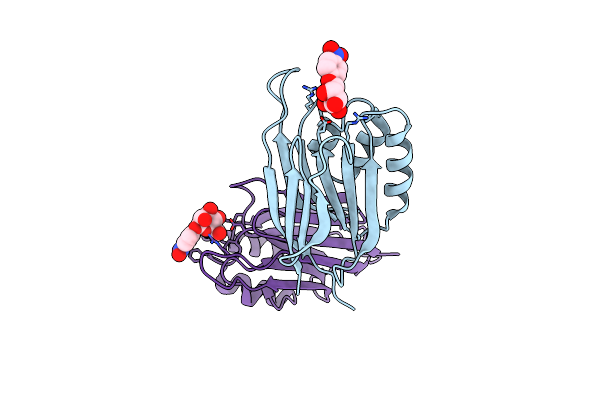 |
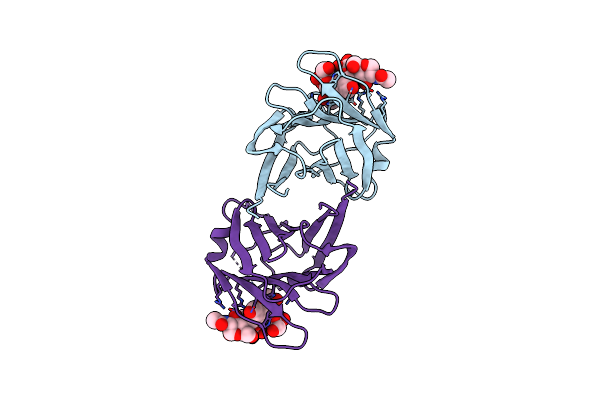
Serial Femtosecond X-Ray Structure Of A Stem Domain Of Human O-Mannose Beta-1,2-N-Acetylglucosaminyltransferase Solved By Se-Sad Using Xfel (Refined Against 13,000 Patterns)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2017-08-30 Classification: SUGAR BINDING PROTEIN Ligands: MBE |
 |
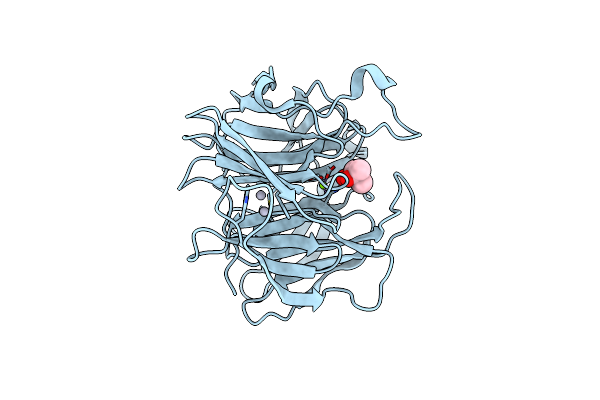
Serial Femtosecond X-Ray Structure Of Agrocybe Cylindracea Galectin With Lactose Solved By Se-Sad Using Xfel (Refined Against 60,000 Patterns)
Organism: Agrocybe cylindracea
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-08-30 Classification: SUGAR BINDING PROTEIN |
 |
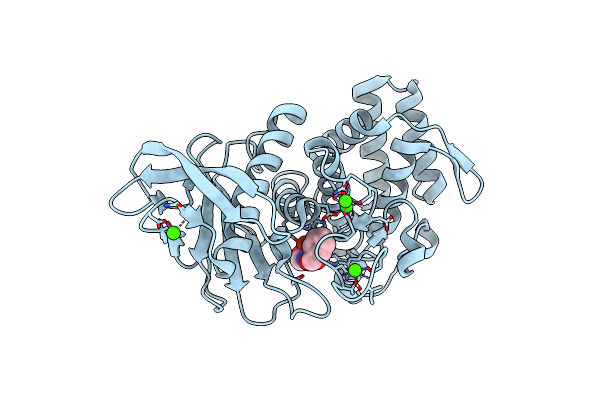
Luciferin-Regenerating Enzyme Solved By Sad Using Xfel (Refined Against 11,000 Patterns)
Organism: Photinus pyralis
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2017-08-30 Classification: HYDROLASE Ligands: HG, MG, MPD |
 |
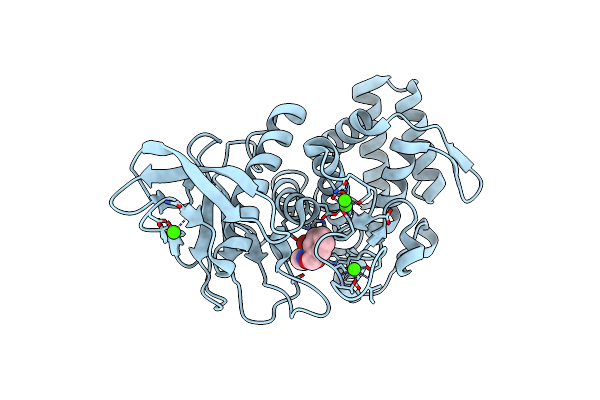
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-08-16 Classification: HYDROLASE Ligands: NX6, ZN, CA |
 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-08-16 Classification: HYDROLASE Ligands: NX6, ZN, CA |
 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-08-16 Classification: HYDROLASE Ligands: ZN, CA |
 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-08-16 Classification: HYDROLASE Ligands: NX6, ZN, CA, PG4 |
 |
Organism: Geobacillus stearothermophilus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-08-16 Classification: HYDROLASE Ligands: NX6, ZN, CA |
 |
Organism: Engyodontium album
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2017-06-07 Classification: HYDROLASE Ligands: CA, NO3 |
 |
Organism: Engyodontium album
Method: X-RAY DIFFRACTION Resolution:0.98 Å Release Date: 2017-06-07 Classification: HYDROLASE Ligands: NO3, GOL, CA |
 |
 |
 |

