Search Count: 18
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2021-10-13 Classification: TRANSCRIPTION Ligands: CL, MG, GOL, NA |
 |
Crystal Structure Of The Bcl6 Btb Domain In Complex With A Hybrid Btb-Binding (Hbp) Peptide
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2021-10-13 Classification: TRANSCRIPTION |
 |
Organism: Streptomyces antibioticus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-11-06 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Streptomyces antibioticus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-11-06 Classification: BIOSYNTHETIC PROTEIN |
 |
Crystal Structure Of Wild-Type Idmh, A Putative Polyketide Cyclase From Streptomyces Antibioticus
Organism: Streptomyces antibioticus
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-11-06 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Staphylococcus aureus
Method: SOLUTION NMR Release Date: 2016-01-27 Classification: TRANSLATION/ANTIBIOTIC RESISTANCE Ligands: ZN |
 |
Direct Visualisation Of Strain-Induced Protein Prost-Translational Modification
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-03-25 Classification: LYASE Ligands: SCN, ACO, MG |
 |
Direct Visualisation Of Strain-Induced Protein Post-Translational Modification
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-03-25 Classification: LYASE Ligands: SCN, ACO, MG |
 |
The Solution Structure Of Monomeric Hepatitis C Virus P7 Yields Potent Inhibitors Of Virion Release
Organism: Hepatitis c virus
Method: SOLUTION NMR Release Date: 2013-09-04 Classification: TRANSPORT PROTEIN |
 |
 |
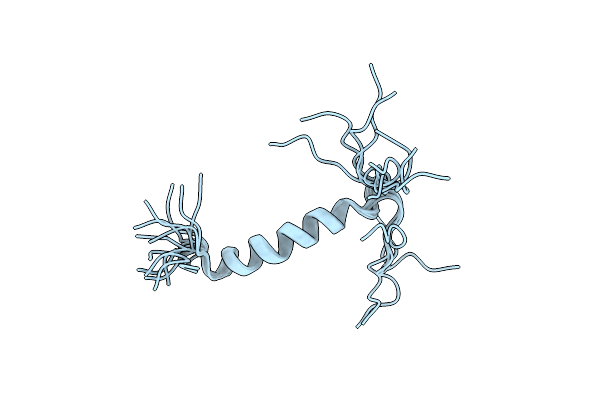 |
Solution Structure Of The Leader Sequence Of The Patellamide Precursor Peptide, Pate1-34
Organism: Prochloron didemni
Method: SOLUTION NMR Release Date: 2010-09-01 Classification: UNKNOWN FUNCTION |
 |
Structure Ensemble Backbone And Side-Chain 1H, 13C, And 15N Chemical Shift Assignments, 1H-15N Rdcs And 1H-1H Noe Restraints For Protein K7 From The Vaccinia Virus
Organism: Vaccinia virus
Method: SOLUTION NMR Release Date: 2008-10-28 Classification: VIRAL PROTEIN |
 |
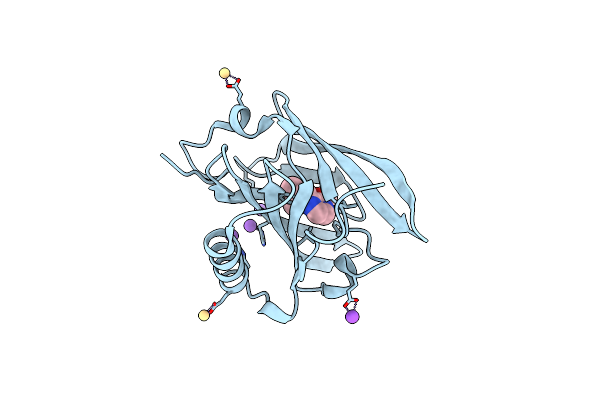
Thermodynamics Of Binding Of 2-Methoxy-3-Isopropylpyrazine And 2-Methoxy-3-Isobutylpyrazine To The Major Urinary Protein
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-02-24 Classification: TRANSPORT PROTEIN Ligands: CD, NA, GOL |
 |
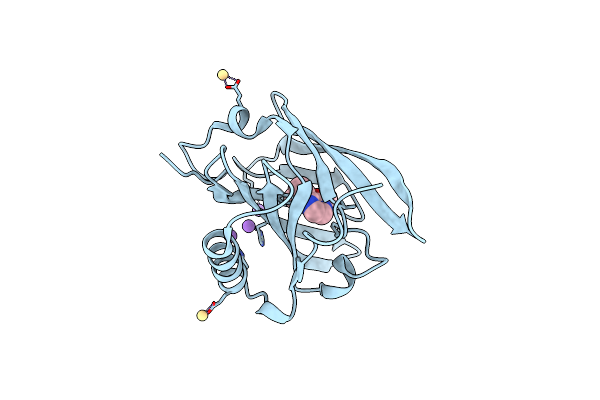
Thermodynamics Of Binding Of 2-Methoxy-3-Isopropylpyrazine And 2-Methoxy-3-Isobutylpyrazine To The Major Urinary Protein
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2004-02-24 Classification: TRANSPORT PROTEIN Ligands: CD, NA, PRZ |
 |
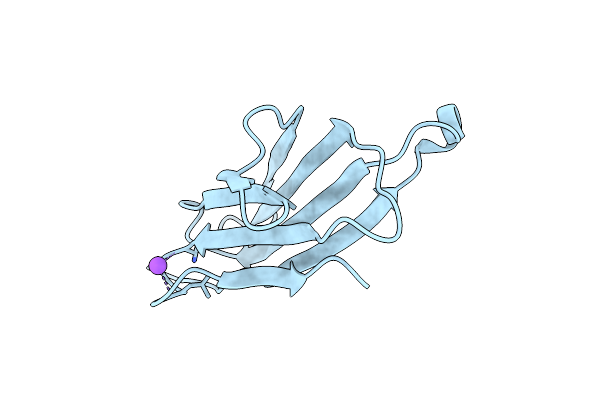
Thermodynamics Of Binding Of 2-Methoxy-3-Isopropylpyrazine And 2-Methoxy-3-Isobutylpyrazine To The Major Urinary Protein
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2004-02-24 Classification: TRANSPORT PROTEIN Ligands: CD, NA, IPZ |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2002-07-31 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of Amicyanin From Paracoccus Versutus (Thiobacillus Versutus)
Organism: Paracoccus versutus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2001-04-11 Classification: ELECTRON TRANSPORT Ligands: CU |

