Search Count: 14
 |
Discovery Of A Novel And Selective Ido-1 Inhibitor Pf-06840003 And Its Characterization As A Potential Clinical Candidate.
Organism: Homo sapiens
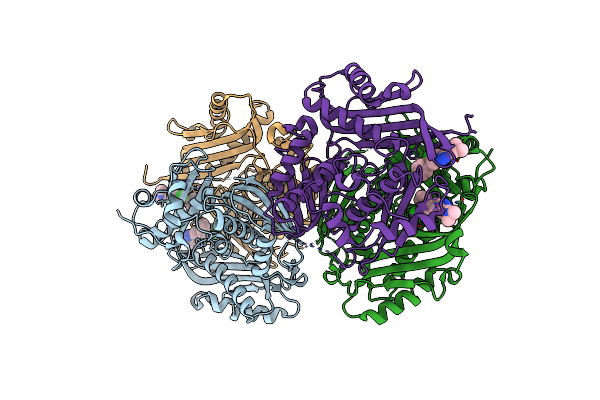
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2017-12-27 Classification: OXIDOREDUCTASE/Inhibitor Ligands: HEM, AOJ |
 |
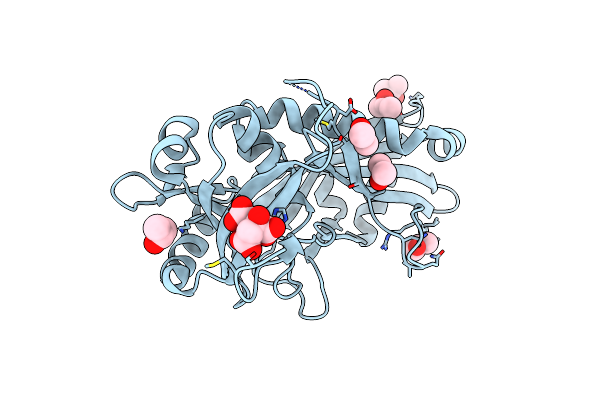
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2017-05-17 Classification: TRANSFERASE Ligands: 8AJ |
 |
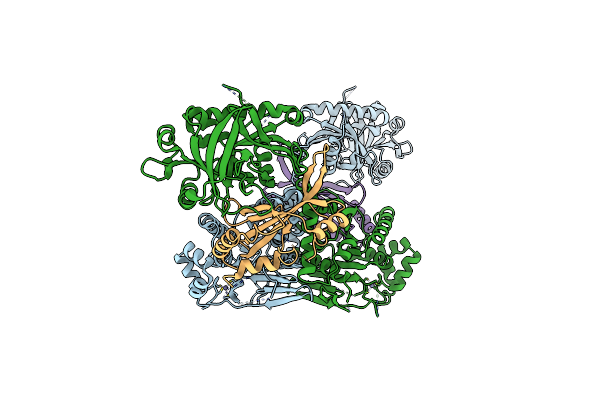
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2012-11-14 Classification: PROTEIN TRANSPORT Ligands: CIT, IPA |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2012-11-14 Classification: PROTEIN TRANSPORT/LIGASE Ligands: ZN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2012-11-14 Classification: PROTEIN TRANSPORT Ligands: ZN |
 |
Crystal Structure Of The Mouse Hoil1-L-Nzf In Complex With Linear Di-Ubiquitin
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2011-12-14 Classification: SIGNALING PROTEIN/METAL BINDING PROTEIN Ligands: ZN |
 |
Crystal Structure Of The Mouse Hoil1-L-Nzf In Complex With Linear Di-Ubiquitin
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-12-14 Classification: SIGNALING PROTEIN/METAL BINDING PROTEIN Ligands: TAM, ZN |
 |
Atg8 Transfer From Atg7 To Atg3: A Distinctive E1-E2 Architecture And Mechanism In The Autophagy Pathway
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2011-11-23 Classification: LIGASE Ligands: ZN |
 |
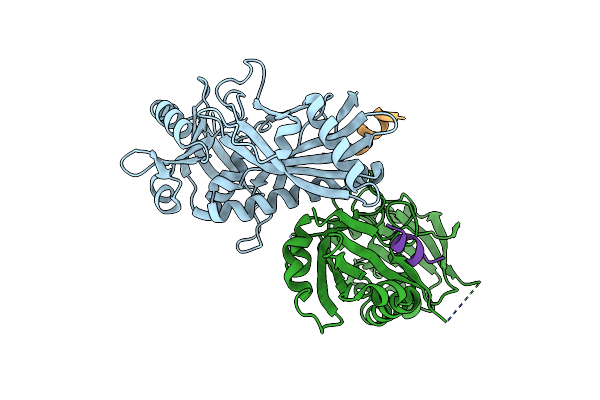
Atg8 Transfer From Atg7 To Atg3: A Distinctive E1-E2 Architecture And Mechanism In The Autophagy Pathway
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2011-11-23 Classification: LIGASE |
 |
Atg8 Transfer From Atg7 To Atg3: A Distinctive E1-E2 Architecture And Mechanism In The Autophagy Pathway
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2011-11-23 Classification: LIGASE |
 |
Atg8 Transfer From Atg7 To Atg3: A Distinctive E1-E2 Architecture And Mechanism In The Autophagy Pathway
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2011-11-23 Classification: LIGASE |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2007-11-20 Classification: MEMBRANE PROTEIN |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2005-07-19 Classification: PROTEIN BINDING |
 |
1.9 Angstrom Crystal Structure Of The Rat Vap-A Msp Homology Domain In Complex With The Rat Orp1 Ffat Motif
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2005-07-19 Classification: PROTEIN BINDING/LIPID BINDING PROTEIN |

