Search Count: 64
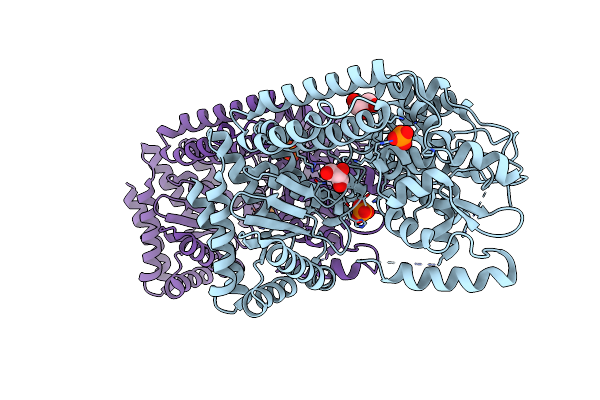 |
Organism: Bacillus sp. d6
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2020-03-04 Classification: HYDROLASE Ligands: GOL, PO4 |
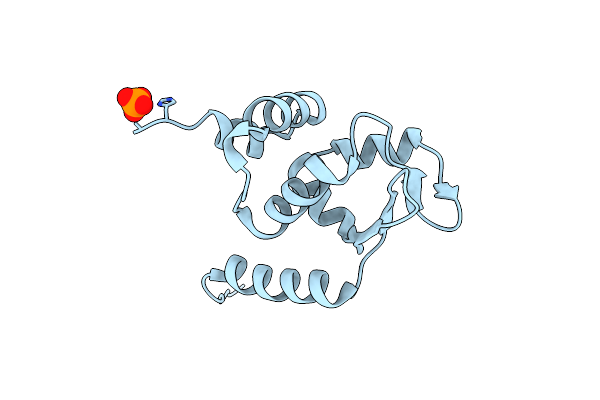 |
Crystal Structure Of Btb-Domain Of Cp190 From D.Melanogaster At High Resolution
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2018-08-29 Classification: TRANSCRIPTION Ligands: PO4 |
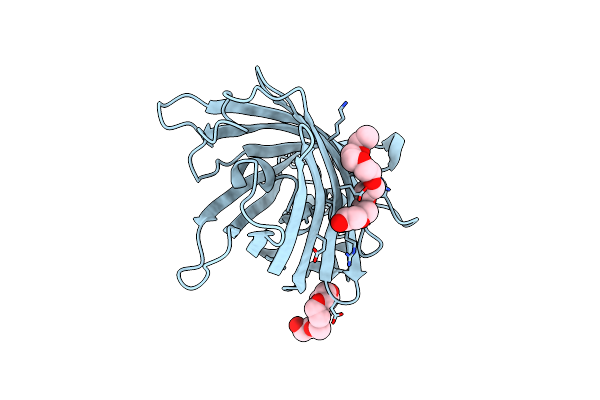 |
Organism: Aequorea victoria
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2018-03-21 Classification: Calcium binding protein Ligands: AE4 |
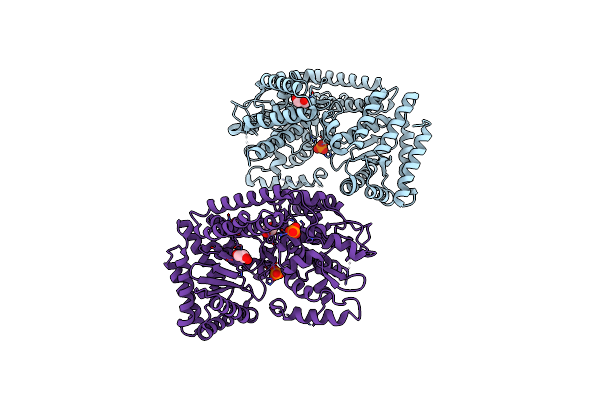 |
The Structure Of D456A Mutant Of Nt.Bspd6I Nicking Endonuclease At 0.24 Nm Resolution .
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-08-16 Classification: HYDROLASE Ligands: GOL, PO4 |
 |
The Structure Of C160S,C508S,C578S Mutant Of Nt.Bspd6I Nicking Endonuclease At 0.185 Nm Resolution .
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-08-02 Classification: HYDROLASE Ligands: GOL, PO4 |
 |
The Structure Of Nt.Bspd6I Nicking Endonuclease With All Cysteines Mutated By Serine Residues At 0.19 Nm Resolution .
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2017-08-02 Classification: HYDROLASE Ligands: PO4 |
 |
Structure Of Lysozyme Labeled With Fluorescein Isothiocyanate (Fitc) At 1.15 Angstroms Resolution
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2017-03-08 Classification: HYDROLASE Ligands: 6B9 |
 |
The 1.5 Angstrom Crystal Structure Of Oxidized (Cuii) Poplar Plastocyanin B At Ph 4.0
Organism: Populus nigra
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-02-13 Classification: ELECTRON TRANSPORT Ligands: CU, SO4, GOL |
 |
The 1.35 Angstrom Crystal Structure Of Reduced (Cui) Poplar Plastocyanin B At Ph 4.0
Organism: Populus nigra
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2013-02-13 Classification: ELECTRON TRANSPORT Ligands: CU1, SO4, GOL, ACT |
 |
The 1.8 Angstrom Crystal Structure Of Oxidized (Cuii) Poplar Plastocyanin B At Ph 6.0
Organism: Populus nigra
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-02-13 Classification: ELECTRON TRANSPORT Ligands: CU, SO4, GOL, ACT |
 |
The 1.54 Angstrom Crystal Structure Of Reduced (Cui) Poplar Plastocyanin B At Ph 6.0
Organism: Populus nigra
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2013-02-13 Classification: ELECTRON TRANSPORT Ligands: CU1, SO4, GOL |
 |
The 1.88 Angstrom Crystal Structure Of Oxidized (Cuii) Poplar Plastocyanin B At Ph 8.0
Organism: Populus nigra
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2013-02-13 Classification: ELECTRON TRANSPORT Ligands: CU, SO4, GOL |
 |
The 1.67 Angstrom Crystal Structure Of Reduced (Cui) Poplar Plastocyanin B At Ph 8.0
Organism: Populus nigra
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2013-02-13 Classification: ELECTRON TRANSPORT Ligands: CU1, GOL |
 |
The 1.08 Angstrom Crystal Structure Of Oxidized (Cuii) Poplar Plastocyanin A At Ph 4.0
Organism: Populus nigra
Method: X-RAY DIFFRACTION Resolution:1.08 Å Release Date: 2013-02-13 Classification: ELECTRON TRANSPORT Ligands: CU, SO4 |
 |
The 1.07 Angstrom Crystal Structure Of Reduced (Cui) Poplar Plastocyanin A At Ph 4.0
Organism: Populus nigra
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2013-02-13 Classification: ELECTRON TRANSPORT Ligands: CU1, SO4 |
 |
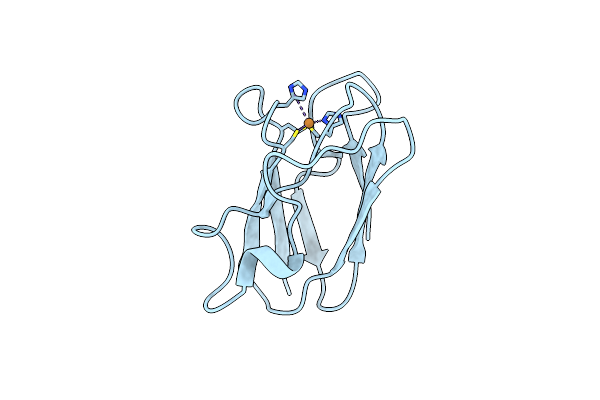
The 1.00 Angstrom Crystal Structure Of Oxidized (Cuii) Poplar Plastocyanin A At Ph 6.0
Organism: Populus nigra
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2013-02-13 Classification: ELECTRON TRANSPORT Ligands: CU |
 |
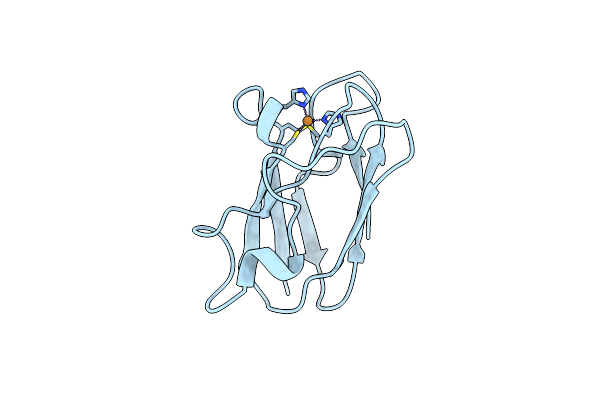
The 1.05 Angstrom Crystal Structure Of Reduced (Cui) Poplar Plastocyanin A At Ph 6.0
Organism: Populus nigra
Method: X-RAY DIFFRACTION Resolution:1.05 Å Release Date: 2013-02-13 Classification: ELECTRON TRANSPORT Ligands: CU1 |
 |
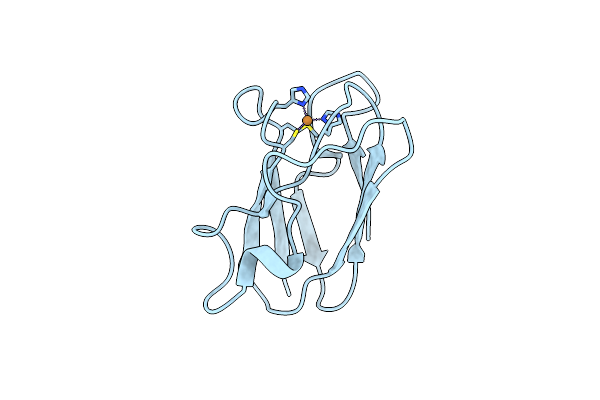
The 1.00 Angstrom Crystal Structure Of Oxidized (Cuii) Poplar Plastocyanin A At Ph 8.0
Organism: Populus nigra
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2013-02-13 Classification: ELECTRON TRANSPORT Ligands: CU |
 |
The 1.06 Angstrom Crystal Structure Of Reduced (Cui) Poplar Plastocyanin A At Ph 8.0
Organism: Populus nigra
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2013-02-13 Classification: ELECTRON TRANSPORT Ligands: CU1 |
 |
Crystal Structure Of Holo D-Serine Dehydratase From Escherichia Coli At 1.55 A Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2012-01-18 Classification: LYASE Ligands: GOL, PLP, K |

