Search Count: 20
 |
Crystal Structure Of The F27G Aim2 Pyrin Domain Mutant And Similarities Of Its Self-Association To Ded/Ded Interactions
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2014-02-19 Classification: SIGNALING PROTEIN, apoptosis |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2012-08-01 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.79 Å Release Date: 2012-08-01 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 0TE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2012-07-25 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2012-07-25 Classification: PROTEIN BINDING Ligands: C2E |
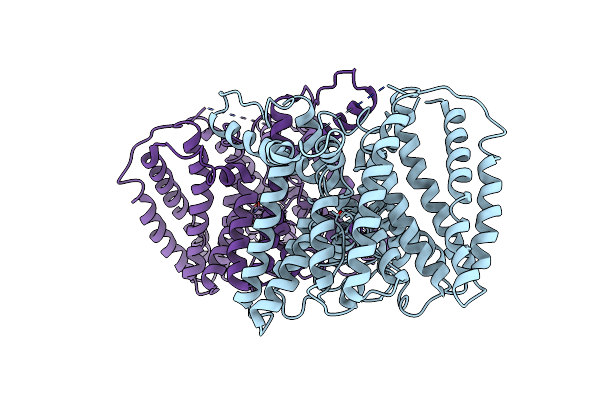 |
Crystal Structure Of The Potassium Transporter Trkh From Vibrio Parahaemolyticus
Organism: Vibrio parahaemolyticus
Method: X-RAY DIFFRACTION Resolution:3.51 Å Release Date: 2011-01-19 Classification: TRANSPORT PROTEIN Ligands: K |
 |
Organism: Mus musculus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:6.80 Å Release Date: 2010-10-13 Classification: APOPTOSIS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.67 Å Release Date: 2010-04-28 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2010-04-28 Classification: SIGNALING PROTEIN Ligands: ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-04-28 Classification: SIGNALING PROTEIN Ligands: ZN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.59 Å Release Date: 2009-03-03 Classification: HYDROLASE Ligands: PO4 |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2008-09-23 Classification: TRANSPORT PROTEIN, oxidoreductase Ligands: NDP, PDN |
 |
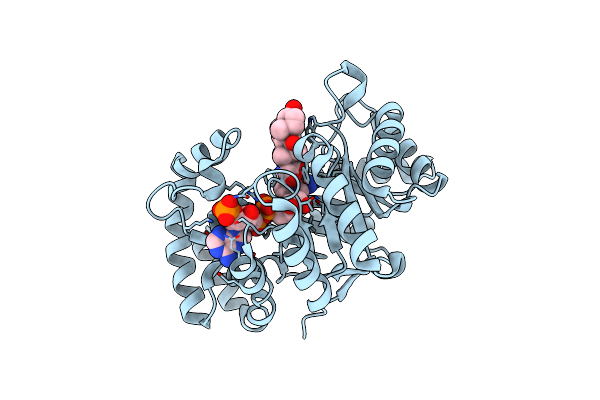
Voltage-Dependent K+ Channel Beta Subunit (W121A) In Complex With Cortisone
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-09-23 Classification: TRANSPORT PROTEIN, oxidoreductase Ligands: NDP, PDN |
 |
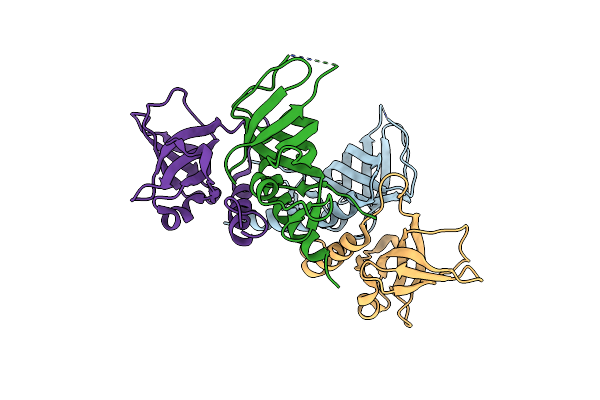
Voltage-Dependent K+ Channel Beta Subunit (I211R) In Complex With Cortisone
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2008-09-23 Classification: TRANSPORT PROTEIN, oxidoreductase Ligands: NDP, PDN |
 |
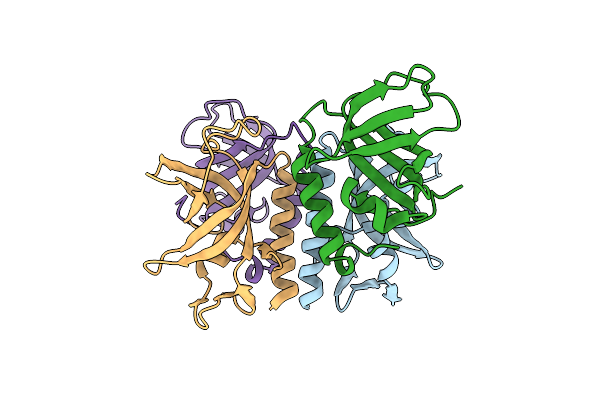
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2007-12-04 Classification: REPLICATION |
 |
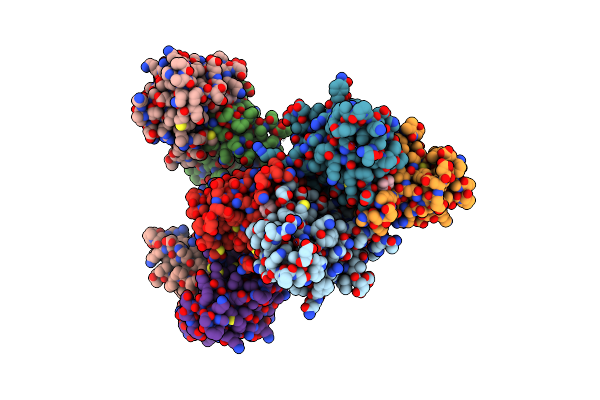
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2007-11-13 Classification: REPLICATION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2007-10-16 Classification: REPLICATION, DNA BINDING PROTEIN Ligands: DIO |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2006-11-28 Classification: HYDROLASE Ligands: MG, ANP, PO4 |
 |
Crystal Structure Of Aspartate-Semialdehyde Dehydrogenase From Vibrio Cholerae El Tor
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2003-03-18 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of Aspartate Semialdehyde Dehydrogenase From Vibrio Cholerae With Nadp And S-Methyl-L-Cysteine Sulfoxide
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2003-01-07 Classification: OXIDOREDUCTASE Ligands: CYS, NDP |

