Search Count: 788
 |
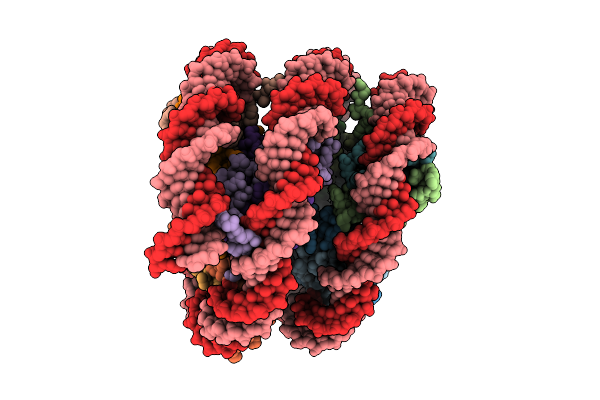
Cryo-Em Structure Of Violaxanthin-Chlorophyll-A-Binding Protein With Red Shifted Chl A (Rvcp) From Trachydiscus Minutus At 2.4 Angstrom
Organism: Trachydiscus minutus
Method: ELECTRON MICROSCOPY Resolution:2.42 Å Release Date: 2025-12-31 Classification: PHOTOSYNTHESIS Ligands: CLA, A1L1F, XAT |
 |
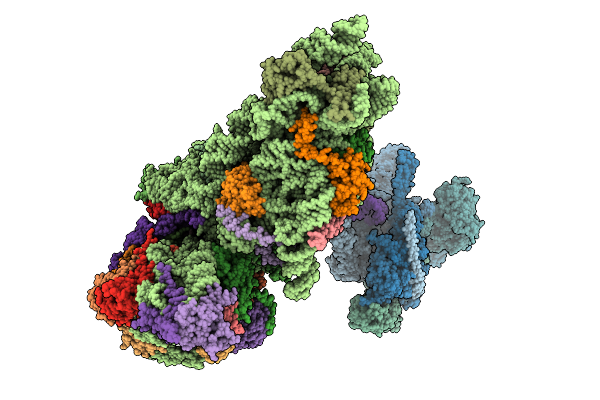
Organism: Xenopus laevis, Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:4.94 Å Release Date: 2025-12-10 Classification: GENE REGULATION |
 |
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG |
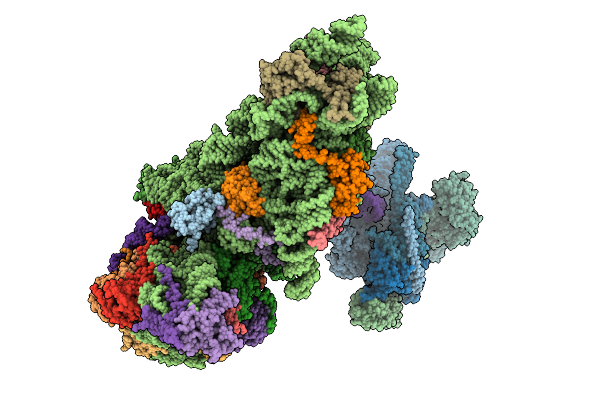 |
Structure Of The Human 40S Ribosome Complexed With Hcv Ires, Eif1A And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG |
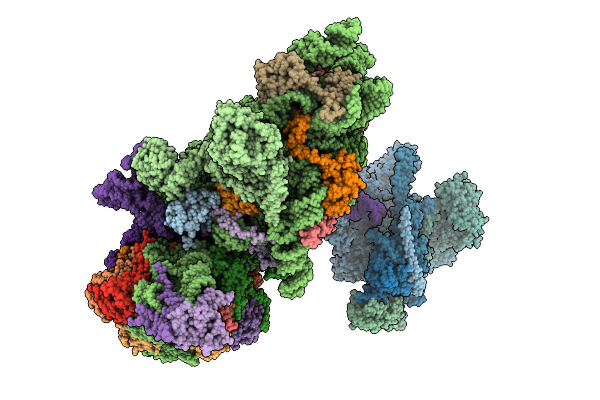 |
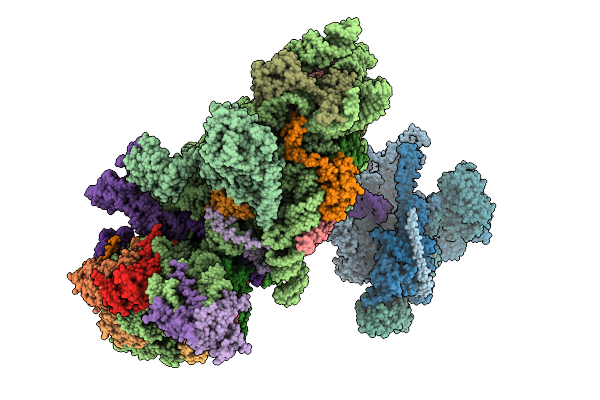
Structure Of The Hcv Ires-Dependent Pre-48S Translation Initiation Complex With Eif1A, Eif5B, And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, GTP, ZN |
 |
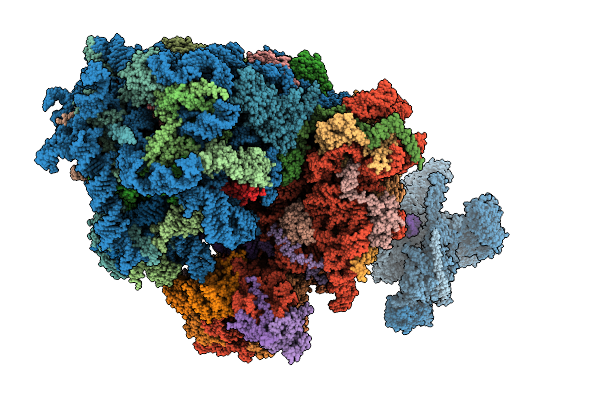
Structure Of The Hcv Ires-Dependent 48S Translation Initiation Complex With Eif5B And Eif3
Organism: Homo sapiens, Hepacivirus hominis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: ZN, MG, GTP |
 |
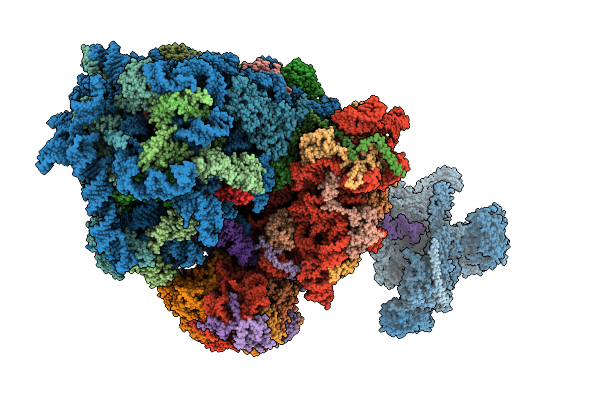
Cryo-Em Structure Of The Hcv Ires-Dependently Initiated Cmv-Stalled 80S Ribosome (Non-Rotated State) In Complexed With Eif3
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, ZN |
 |
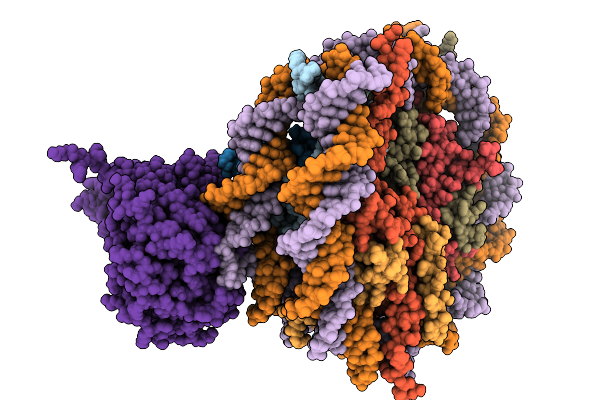
Cryo-Em Structure Of The Hcv Ires-Dependently Initiated Cmv-Stalled 80S Ribosome (Rotated State) In Complexed With Eif3
Organism: Hepacivirus hominis, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: RIBOSOME Ligands: MG, ZN |
 |
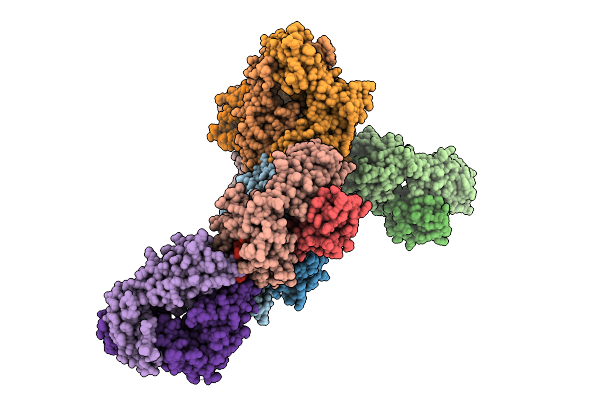
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: DNA BINDING PROTEIN |
 |
Crystal Structure Of Leminorella Grimontii Gatc In The Presence Of D-Xylose
Organism: Leminorella grimontii
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2025-10-15 Classification: TRANSPORT PROTEIN Ligands: OLC |
 |
Human T Cell Receptor (Trav24*01/Trbv27*01) In Complex With Hla-C*1202 And Iy11 Peptide
Organism: Homo sapiens, Human immunodeficiency virus type 1 (z2/cdc-z34 isolate)
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2025-09-17 Classification: IMMUNE SYSTEM |
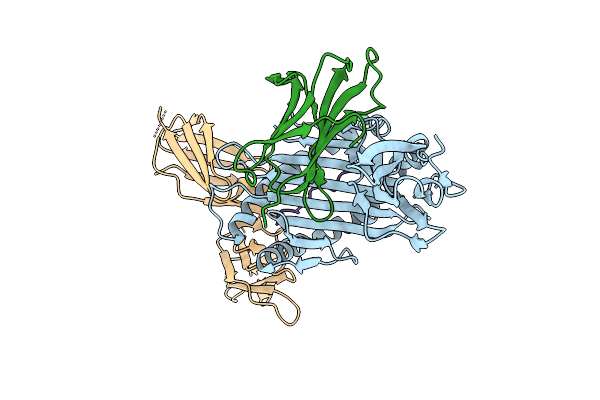 |
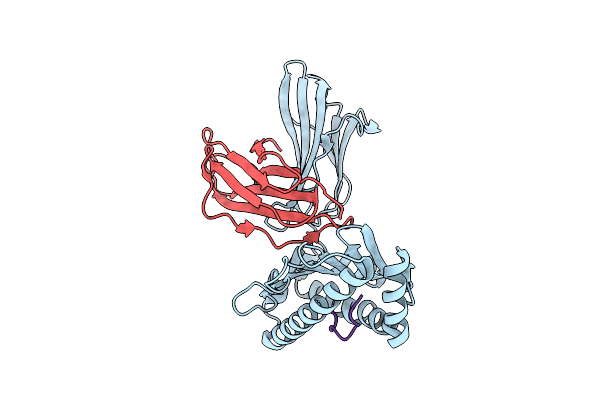
Killer Cell Immunoglobulin-Like Receptor 2Dl2 In Complex With Hla-C*1202 And Iy10 Peptide
Organism: Homo sapiens, Human immunodeficiency virus type 1 (z2/cdc-z34 isolate)
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-09-17 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Human immunodeficiency virus type 1 (z2/cdc-z34 isolate)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-09-17 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Human immunodeficiency virus type 1 (z2/cdc-z34 isolate)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-09-17 Classification: IMMUNE SYSTEM |
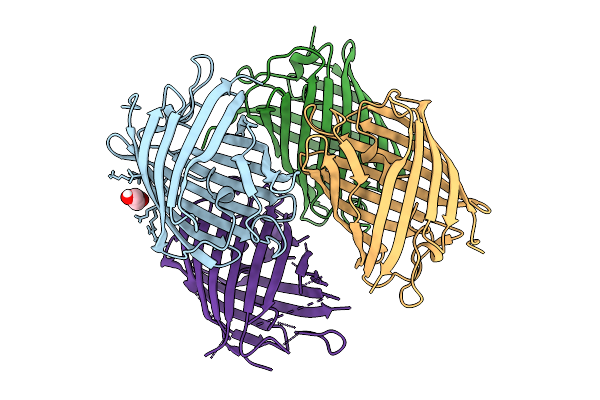 |
X-Ray Crystal Structure Of Kohinoor Reversibly Switchable Fluorescent Protein
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2025-09-03 Classification: FLUORESCENT PROTEIN Ligands: PEG |
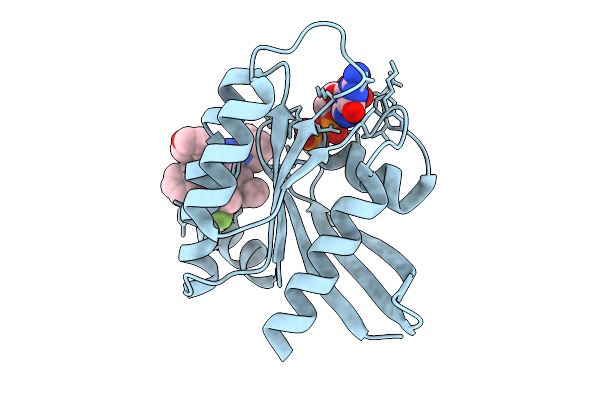 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2025-09-03 Classification: HYDROLASE Ligands: GDP, MG, A1L65 |
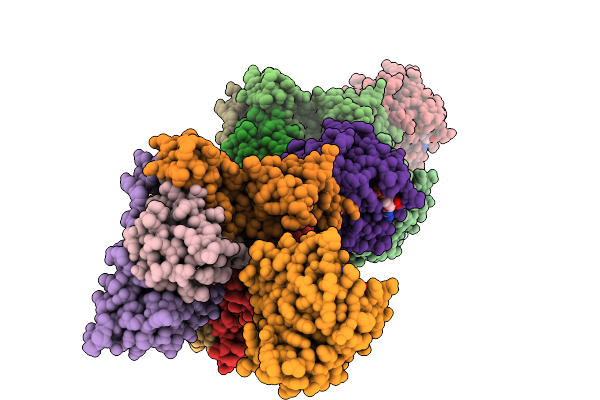 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.18 Å Release Date: 2025-09-03 Classification: HYDROLASE Ligands: A1L66, GDP, MG |
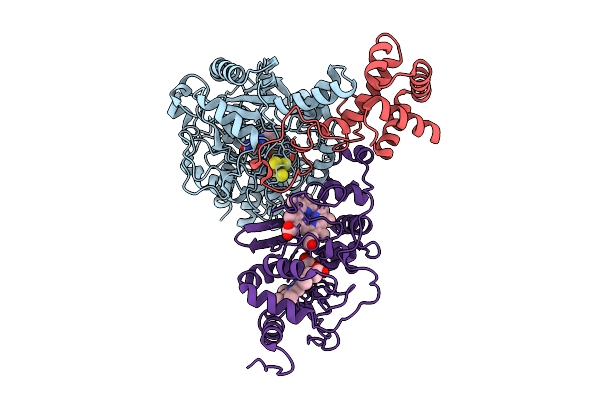 |
Cryo-Em Structure Of Fructose Dehydrogenase Variant From Gluconobacter Japonicus Truncating Heme 1C And C-Terminal Hydrophobic Regions
Organism: Gluconobacter japonicus
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: FAD, F3S, HEC |
 |
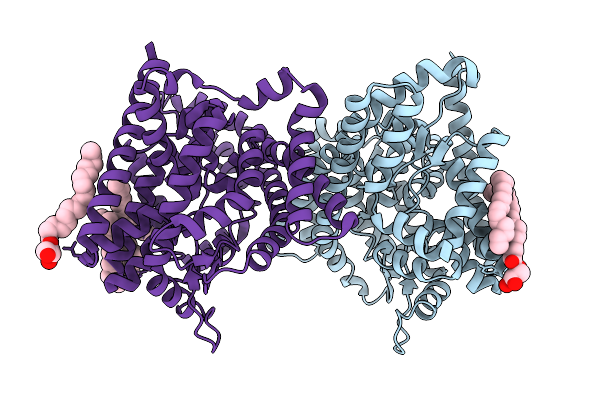
X-Ray Structure Of Cytochrome P450 Olet From Lacicoccus Alkaliphilus In Complex With Icosanoic Acid
Organism: Lacicoccus alkaliphilus dsm 16010
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: HEM, DCR, GOL |
 |
Organism: Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-07-16 Classification: BIOSYNTHETIC PROTEIN Ligands: IPA, CL |

