Search Count: 34
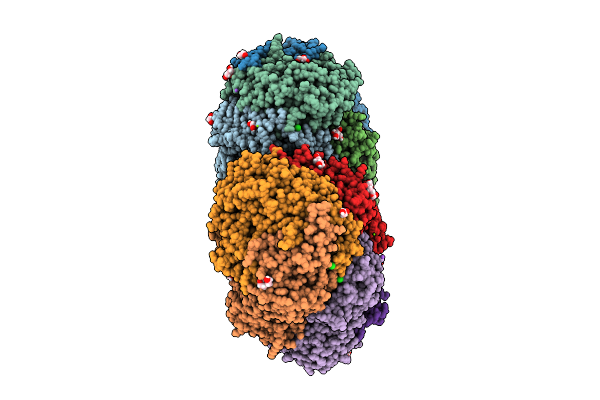 |
Krypton-Pressurized Methyl-Coenzyme M Reductase Of An Anme-2C Isolated From A Microbial Enrichment
Organism: Candidatus methanogasteraceae archaeon
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: KR, MG, PGE, EDO, CL, TP7, F43, PEG, GOL, COM, NA, K |
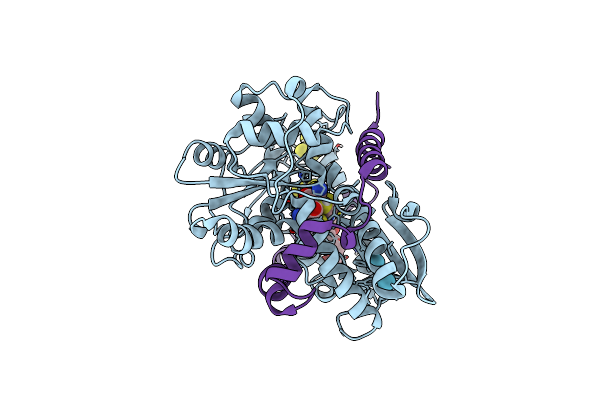 |
Desulfovibrio Desulfuricans [Fefe] Hydrogenase In Hinact State Derivatized With Krypton
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2025-02-19 Classification: OXIDOREDUCTASE Ligands: KR, SF4, TOE, LFH |
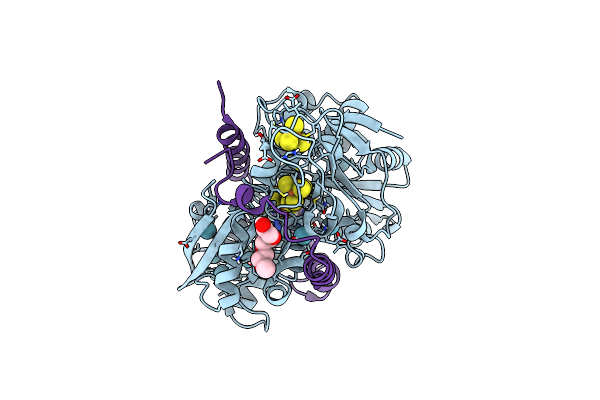 |
Desulfovibrio Desulfuricans [Fefe] Hydrogenase C178A Mutant In Hinact-Like State Derivatized With Krypton
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2025-02-19 Classification: OXIDOREDUCTASE Ligands: KR, 402, SF4, CYN, TOE |
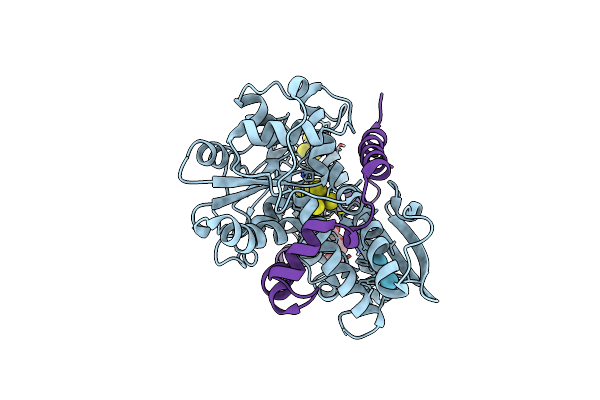 |
Desulfovibrio Desulfuricans [Fefe] Hydrogenase In The Apo Form Derivatized With Krypton
Organism: Desulfovibrio desulfuricans
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: KR, SF4, PGE |
 |
Crystal Structure Of A Flavodiiron Protein D52K Mutant From Escherichia Coli Pressurized With Krypton Gas
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: FMN, FE, O, KR, OXY |
 |
Organism: Halobacterium salinarum (strain atcc 700922 / jcm 11081 / nrc-1)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-04-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA, OLA, HEX, SO4, KR |
 |
Organism: Dokdonia eikasta
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-04-27 Classification: MEMBRANE PROTEIN Ligands: LFA, HEX, NA, KR, RET |
 |
Organism: Candidatus actinomarina minuta
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-04-27 Classification: MEMBRANE PROTEIN Ligands: RET, LFA, HEX, KR |
 |
Aerobic Crystal Structure Of F420H2-Oxidase From Methanothermococcus Thermolithotrophicus At 1.8A Resolution Under 125 Bars Of Krypton
Organism: Methanothermococcus thermolithotrophicus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-11-25 Classification: OXIDOREDUCTASE Ligands: HEZ, FEO, KR, FMN, CL |
 |
Structure Of Hyperstable Haloalkane Dehalogenase Variant Dhaa115 Prepared By The 'Soak-And-Freeze' Method Under 150 Bar Of Krypton Pressure
Organism: Rhodococcus rhodochrous
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2020-11-18 Classification: HYDROLASE Ligands: KR, SCN, B3P, GOL |
 |
Structure Of [Nifese] Hydrogenase From Desulfovibrio Vulgaris Hildenborough Pressurized With Krypton Gas - Structure Wtkr1
Organism: Desulfovibrio vulgaris (strain hildenborough / atcc 29579 / dsm 644 / ncimb 8303)
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2020-09-09 Classification: OXIDOREDUCTASE Ligands: SF4, 6ML, KR, FCO, NI, FE2, H2S, CL |
 |
Structure Of [Nifese] Hydrogenase G491A Variant From Desulfovibrio Vulgaris Hildenborough Pressurized With Krypton Gas - Structure G491A-Kr
Organism: Desulfovibrio vulgaris (strain hildenborough / atcc 29579 / dsm 644 / ncimb 8303)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-09-09 Classification: OXIDOREDUCTASE Ligands: SF4, 6ML, KR, FCO, NI, FE2, H2S, CL |
 |
Structure Of The Trna-Monooxygenase Enzyme Miae Frozen Under 140 Bar Of Krypton Using The Soak And Freeze Methodology
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-09-02 Classification: OXIDOREDUCTASE Ligands: FE, CL, CA, TRS, PEG, PG4, KR, EDO |
 |
Organism: Aspergillus flavus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-05-13 Classification: OXIDOREDUCTASE Ligands: MPD, ACT, AZA, KR, NA |
 |
Organism: Aspergillus flavus
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2019-12-18 Classification: OXIDOREDUCTASE Ligands: MPD, AZA, ACT, KR, NA |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-11-29 Classification: OXYGEN TRANSPORT Ligands: SO4, HEM, KR |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-11-15 Classification: OXYGEN TRANSPORT Ligands: HEM, SO4, KR |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-11-15 Classification: OXYGEN STORAGE Ligands: HEM, SO4, KR |
 |
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2017-08-16 Classification: HYDROLASE Ligands: ZN, CA, GOL, 7GR, DMS, KR |
 |
Structure Of Thermolysin Prepared By The 'Soak-And-Freeze' Method Under 100 Bar Of Krypton Pressure
Organism: Bacillus thermoproteolyticus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-10-26 Classification: HYDROLASE Ligands: VAL, LYS, ZN, KR, CA |

