Search Count: 4,798
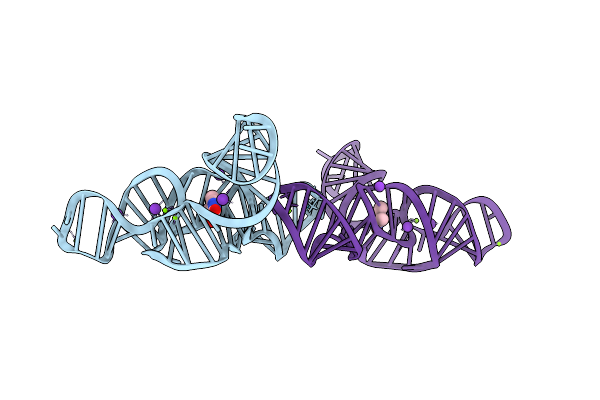 |
Structure Of The Bacillus Subtilis Yjdf Riboswitch Aptamer Domain In Complex With Lumichrome
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: RNA Ligands: MG, K, LUM |
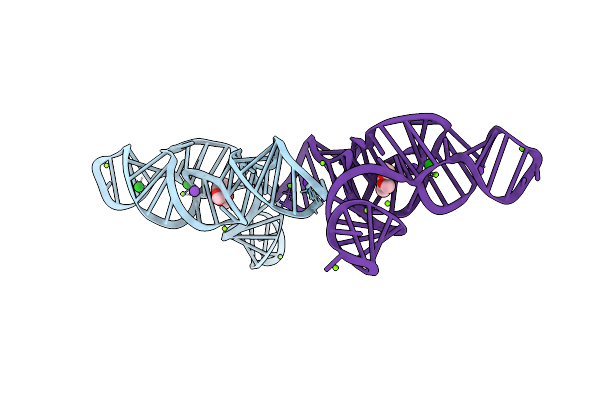 |
Structure Of The Bacillus Subtilis Yjdf Riboswitch Aptamer Domain In Complex With Chelerythrine
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: RNA Ligands: CTI, BA, K, MG |
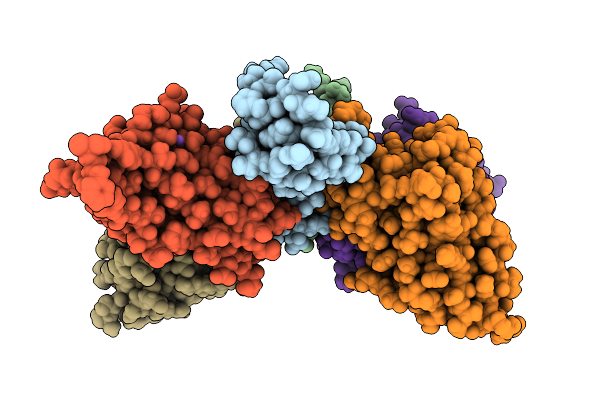 |
Organism: Homo sapiens, Naja kaouthia
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: TOXIN Ligands: GOL, CL, K, NA, SO4 |
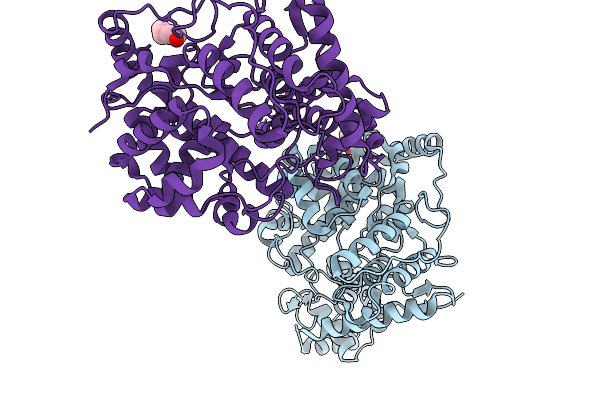 |
Organism: Orthonairovirus songlingense
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: VIRAL PROTEIN Ligands: PEG, CL, K |
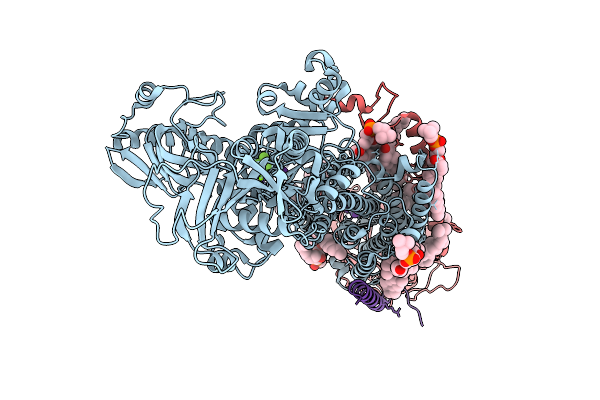 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: ALF, MG, K, PCW, CLR |
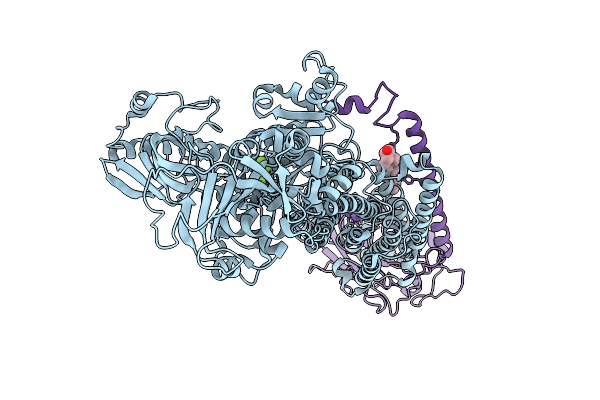 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: MEMBRANE PROTEIN Ligands: ALF, MG, K, CLR |
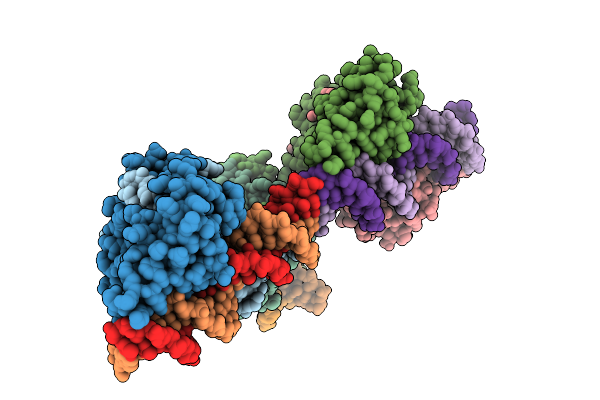 |
Organism: Clostridioides difficile
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSCRIPTION Ligands: K |
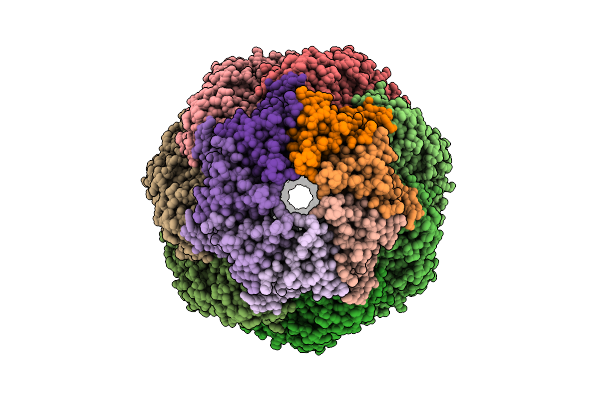 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: CHAPERONE Ligands: ATP, MG, K |
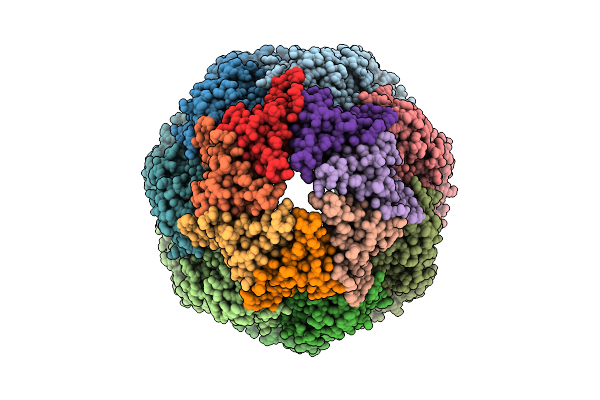 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: CHAPERONE Ligands: ATP, MG, K |
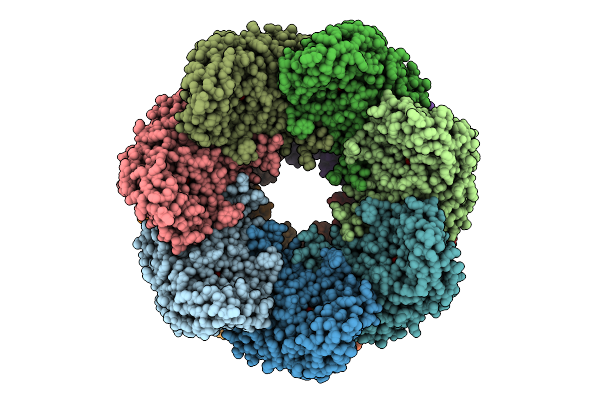 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: CHAPERONE Ligands: ATP, MG, K |
 |
K115 Acetylated Human Muscle Pyruvate Kinase, Isoform M2 (Pkm2), In Complex With Fbp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSFERASE Ligands: FBP, EDO, SIN, GOL, K |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: TRANSFERASE Ligands: GOL, EDO, MG, TRS, SIN, K |
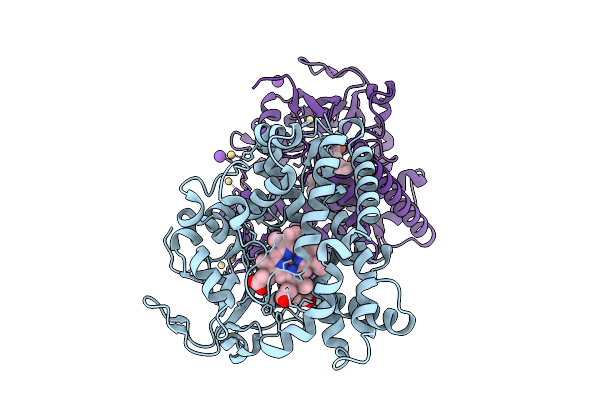 |
Crystal Structure Of The Cyp154C5 F92A/R114A/T248D/E282A Variant From Nocardia Farcinica
Organism: Nocardia farcinica ifm 10152
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: OXIDOREDUCTASE Ligands: TES, HEM, CD, CL, K, GOL |
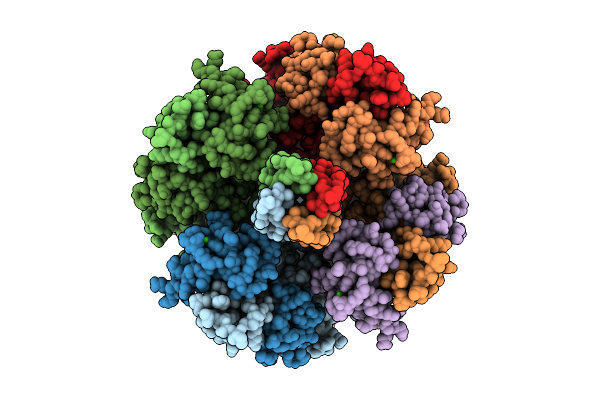 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: PT5, K, CA |
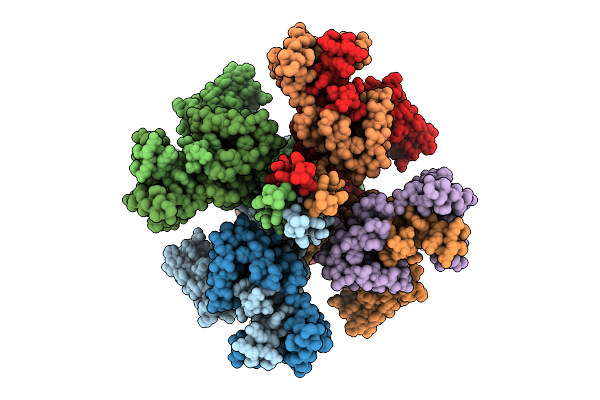 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: MEMBRANE PROTEIN Ligands: PIO, K |
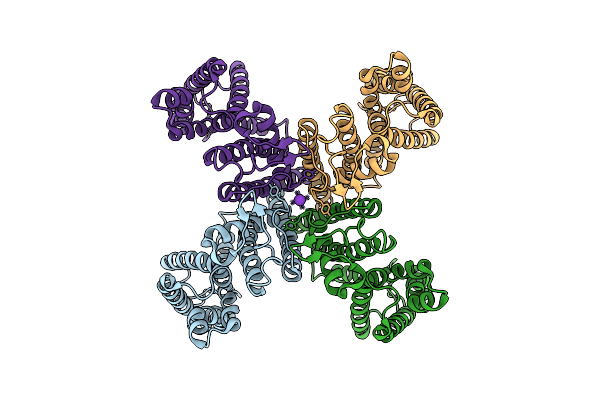 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PLANT PROTEIN Ligands: K |
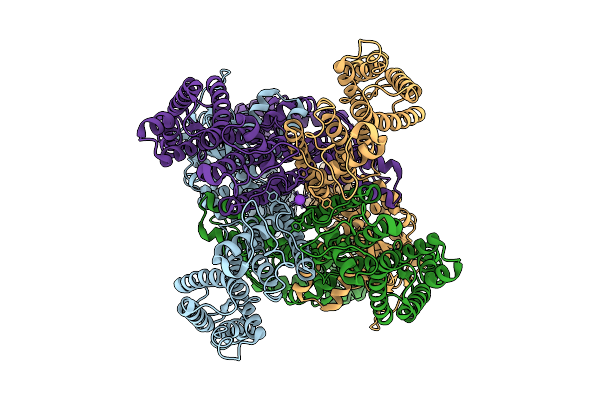 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PROTON TRANSPORT Ligands: K |
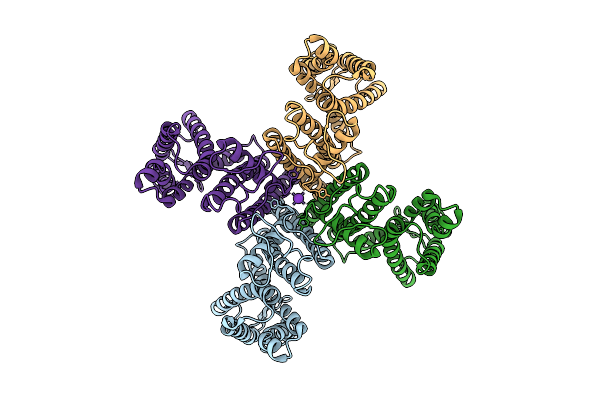 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: PROTON TRANSPORT Ligands: K |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: RIBOSOME Ligands: MG, K, G34, VAL, NAD, SPD, ZN, FES, ATP, GDP |
 |
Organism: Aspergillus fumigatus af293
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: BIOSYNTHETIC PROTEIN Ligands: A1BDC, ACY, TRS, PEG, EDO, NA, CL, K |

