Search Count: 26
 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase From Amycolatopsis Mediterranei (Amgh128_I)
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: GOL, NA |
 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase From Amycolatopsis Mediterranei (Amgh128_I) In Complex With Laminaritriose
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-05-20 Classification: HYDROLASE |
 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase (E199A Mutant) From Amycolatopsis Mediterranei (Amgh128_I) In Complex With Laminaripentaose
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: BGC, ZN, PEG |
 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase (E102A Mutant) From Amycolatopsis Mediterranei (Amgh128_I) In Complex With Laminaripentaose
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of A Gh128 (Subgroup I) Endo-Beta-1,3-Glucanase (E102A Mutant) From Amycolatopsis Mediterranei (Amgh128_I) In Complex With Laminaritriose And Laminaribiose
Organism: Amycolatopsis mediterranei
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: ZN, PEG |
 |
Crystal Structure Of A Gh128 (Subgroup Ii) Endo-Beta-1,3-Glucanase From Pseudomonas Viridiflava (Pvgh128_Ii)
Organism: Pseudomonas viridiflava
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Crystal Structure Of A Gh128 (Subgroup Ii) Endo-Beta-1,3-Glucanase From Pseudomonas Viridiflava (Pvgh128_Ii) In Complex With Laminaritriose
Organism: Pseudomonas viridiflava
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: SO4 |
 |
Crystal Structure Of A Gh128 (Subgroup Ii) Endo-Beta-1,3-Glucanase From Sorangium Cellulosum (Scgh128_Ii)
Organism: Sorangium cellulosum so ce56
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: TRS, EPE, CA, CL |
 |
Crystal Structure Of A Gh128 (Subgroup Iii) Curdlan-Specific Exo-Beta-1,3-Glucanase From Blastomyces Gilchristii (Bggh128_Iii)
Organism: Blastomyces gilchristii
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-05-20 Classification: HYDROLASE |
 |
Crystal Structure Of A Gh128 (Subgroup Iii) Curdlan-Specific Exo-Beta-1,3-Glucanase From Blastomyces Gilchristii (Bggh128_Iii) In Complex With Glucose
Organism: Blastomyces gilchristii
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: BGC |
 |
Crystal Structure Of A Gh128 (Subgroup Iii) Curdlan-Specific Exo-Beta-1,3-Glucanase From Blastomyces Gilchristii (Bggh128_Iii) In Complex With Laminaribiose At -2 And -1 Subsites
Organism: Blastomyces gilchristii (strain slh14081)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-05-20 Classification: HYDROLASE |
 |
Crystal Structure Of A Gh128 (Subgroup Iii) Curdlan-Specific Exo-Beta-1,3-Glucanase From Blastomyces Gilchristii (Bggh128_Iii) In Complex With Laminaribiose At -3 And -2 Subsites
Organism: Blastomyces gilchristii
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: BGC |
 |
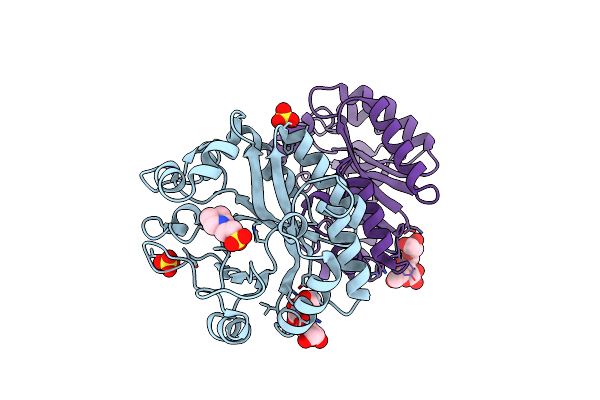
Crystal Structure Of A Gh128 (Subgroup Iv) Endo-Beta-1,3-Glucanase From Lentinula Edodes (Legh128_Iv)
Organism: Lentinula edodes
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: MES, CL, SO4 |
 |
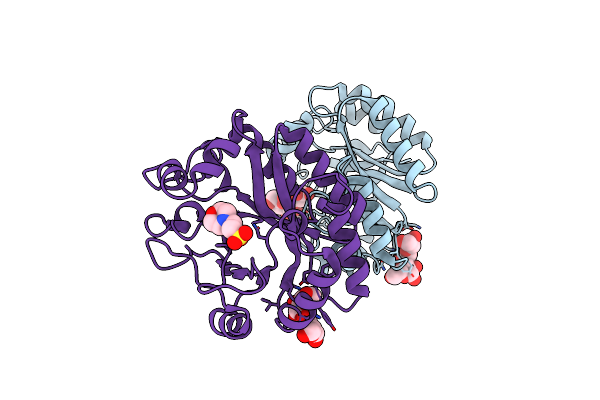
Crystal Structure Of A Gh128 (Subgroup Iv) Endo-Beta-1,3-Glucanase From Lentinula Edodes (Legh128_Iv) With Laminaribiose At The Surface-Binding Site
Organism: Lentinula edodes
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: CL, MES, SO4 |
 |
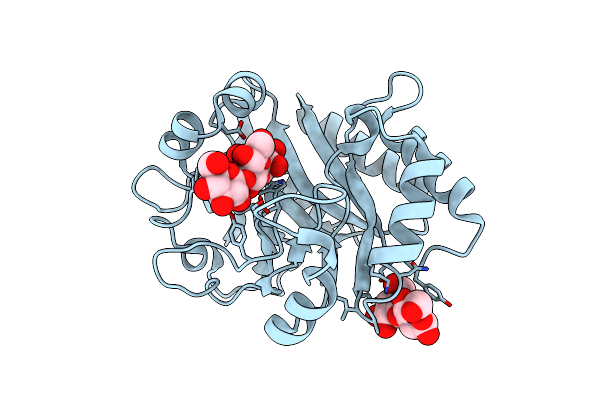
Crystal Structure (C2 Form) Of A Gh128 (Subgroup Iv) Endo-Beta-1,3-Glucanase From Lentinula Edodes (Legh128_Iv) In Complex With Laminaritriose
Organism: Lentinula edodes
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: CL, MES |
 |
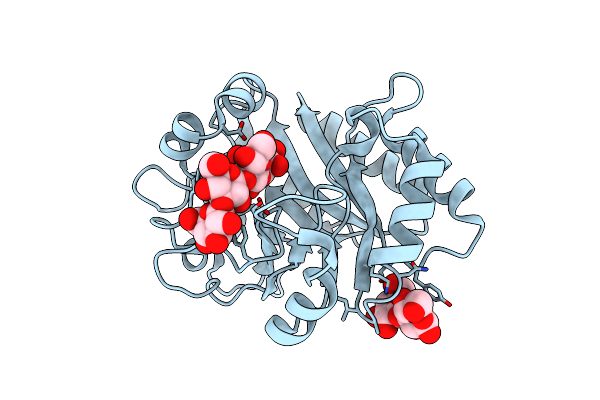
Crystal Structure (P21 Form) Of A Gh128 (Subgroup Iv) Endo-Beta-1,3-Glucanase From Lentinula Edodes (Legh128_Iv) In Complex With Laminaritriose
Organism: Lentinula edodes
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: CL |
 |
Crystal Structure Of A Gh128 (Subgroup Iv) Endo-Beta-1,3-Glucanase From Lentinula Edodes (Legh128_Iv) In Complex With Laminaritetraose
Organism: Lentinula edodes
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: CL |
 |
Crystal Structure Of A Gh128 (Subgroup V) Exo-Beta-1,3-Glucanase From Cryptococcus Neoformans (Cngh128_V)
Organism: Cryptococcus neoformans
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: K |
 |
Crystal Structure Of A Gh128 (Subgroup Vi) Exo-Beta-1,3-Glucanase From Aureobasidium Namibiae (Angh128_Vi)
Organism: Aureobasidium namibiae cbs 147.97
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-05-20 Classification: HYDROLASE Ligands: GOL |
 |
Crystal Structure Of A Gh128 (Subgroup Vi) Exo-Beta-1,3-Glucanase From Aureobasidium Namibiae (Angh128_Vi) In Complex With Laminaritriose
Organism: Aureobasidium namibiae cbs 147.97
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2020-05-20 Classification: HYDROLASE |

