Search Count: 9
 |
Organism: Gluconobacter oxydans
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-03-08 Classification: OXIDOREDUCTASE |
 |
Crystal Structure Of D-Galactose-6-Phosphate Isomerase In A Substrate-Free Form
Organism: Lactobacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2013-11-27 Classification: ISOMERASE |
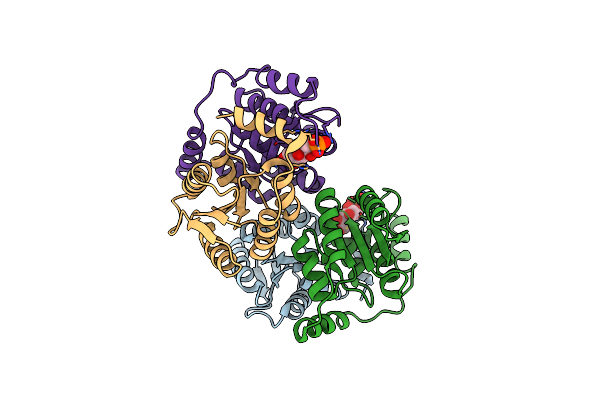 |
Crystal Structure Of D-Galactose-6-Phosphate Isomerase In Complex With D-Tagatose-6-Phosphate
Organism: Lactobacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2013-11-27 Classification: ISOMERASE Ligands: TG6 |
 |
Crystal Structure Of D-Galactose-6-Phosphate Isomerase In Complex With D-Psicose
Organism: Lactobacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2013-11-27 Classification: ISOMERASE Ligands: PSJ |
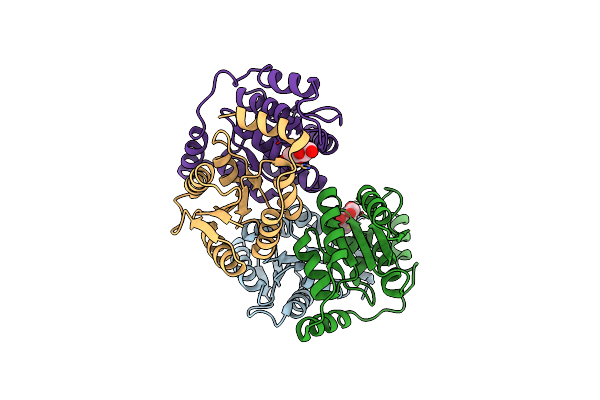 |
Crystal Structure Of D-Galactose-6-Phosphate Isomerase In Complex With D-Ribulose
Organism: Lactobacillus rhamnosus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2013-11-27 Classification: ISOMERASE Ligands: RBL |
 |
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2011-08-10 Classification: TOXOFLAVIN BINDING PROTEIN Ligands: MN |
 |
Crystal Structure Of Toxoflavin-Degrading Enzyme In Complex With Toxoflavin
Organism: Paenibacillus polymyxa
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-08-10 Classification: TOXOFLAVIN BINDING PROTEIN Ligands: MN, TOF |
 |
Organism: Bacillus sp. snu-7
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-12-26 Classification: LYASE Ligands: 2PO |
 |
Crystal Structure Of Inulin Fructotransferase In The Presence Of Di-Fructose
Organism: Bacillus sp. snu-7
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2006-12-26 Classification: LYASE Ligands: 2PO |

