Search Count: 11
 |
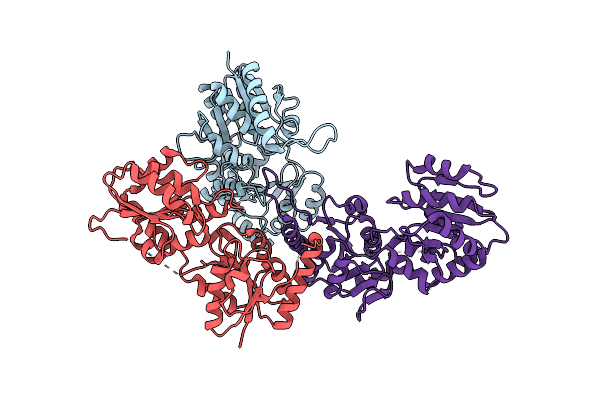
Structure Of Udp-N-Acetylmuramoyl-L-Alanyl-D-Glutamate--2, 6-Diaminopimelate Ligase
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2021-07-28 Classification: LIGASE |
 |
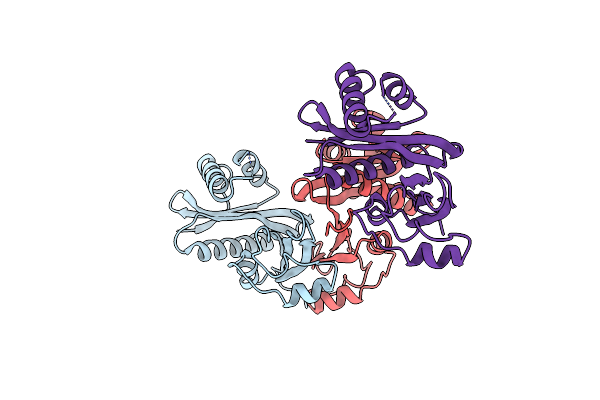
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2021-07-14 Classification: TRANSFERASE |
 |
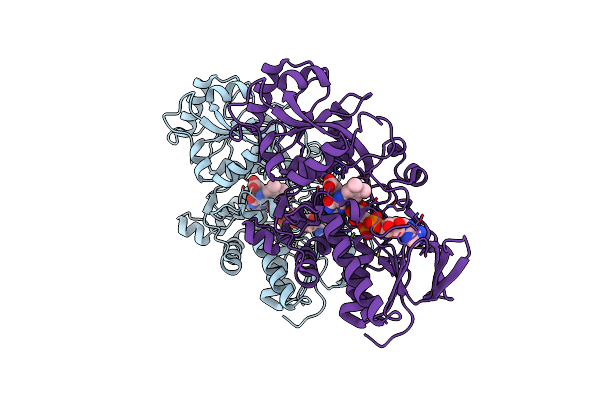
Crystal Structure Of The Aminoglycoside 6'-N-Acetyltransferase From Enterococcus Faecium
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-08-12 Classification: TRANSFERASE |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2020-07-08 Classification: OXIDOREDUCTASE Ligands: FAD, SIN |
 |
Apo Crystal Structure Of Class I Type B Peptide Deformylase From Acinetobacter Baumannii
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-02-12 Classification: HYDROLASE Ligands: ZN, GOL |
 |
Met-Ala-Ser Bound Crystal Structure Of Class I Type B Peptide Deformylase From Acinetobacter Baumannii
Organism: Acinetobacter baumannii, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2020-02-12 Classification: HYDROLASE Ligands: ZN |
 |
K3U Bound Crystal Structure Of Class I Type B Peptide Deformylase From Acinetobacter Baumannii
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-02-12 Classification: HYDROLASE Ligands: ZN, K3U |
 |
Crystal Structure Of The Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, At 1.57 Angstrom
Organism: Acetabularia acetabulum
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2015-08-26 Classification: PROTON TRANSPORT Ligands: RET, OLB, D12, D10, OCT, C14 |
 |
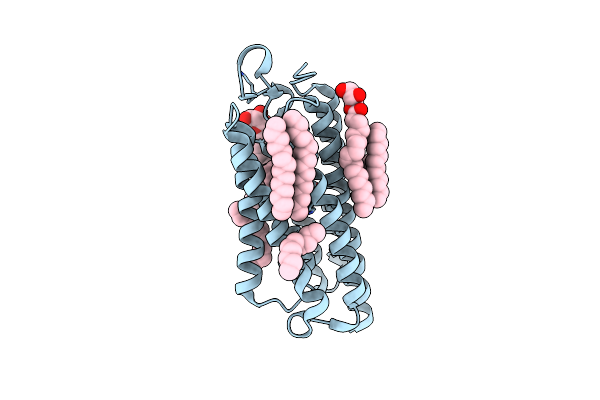
Crystal Structure Of The Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, At 1.52 Angstrom
Organism: Acetabularia acetabulum
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2015-08-26 Classification: PROTON TRANSPORT Ligands: RET, OLB, D12, C14, R16, D10 |
 |
Crystal Structure Of The Cell-Free Synthesized Membrane Protein, Acetabularia Rhodopsin I, At 1.80 Angstrom
Organism: Acetabularia acetabulum
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-08-26 Classification: PROTON TRANSPORT Ligands: RET, OLB, C14, D10, R16, D12 |
 |
Structure Of Anabaena Sensory Rhodopsin Determined By Solid State Nmr Spectroscopy
Organism: Nostoc sp. pcc 7120
Method: SOLID-STATE NMR Release Date: 2013-08-21 Classification: MEMBRANE PROTEIN Ligands: RET |

