Search Count: 32
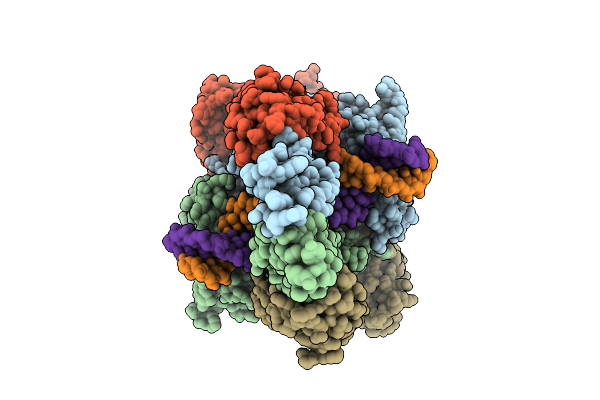 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: HYDROLASE Ligands: MG, ADP, BEF, MN |
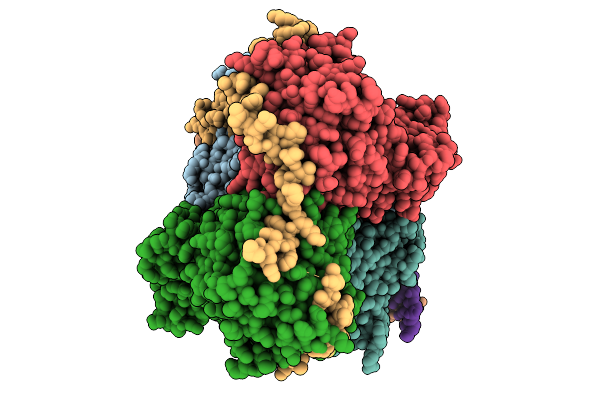 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: HYDROLASE Ligands: MG, ADP, BEF, MN |
 |
Cryo-Em Structure Of Human Mre11-Rad50-Nbs1 (Mrn) Complex Bound To Dna And Telomeric Factor Trf2 Fragment (438-542)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: HYDROLASE Ligands: MG, ADP, BEF, MN |
 |
Cryo-Em Structure Of Human Mre11-Rad50 (Mr) Complex Bound To Dna And Telomeric Factor Trf2 Fragment (438-542)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: HYDROLASE Ligands: ADP, MG, BEF, MN |
 |
Cryo-Em Structure Of Human Mre11-Rad50-Nbs1 (Mrn) Complex Bound To Dna And Telomeric Factor Trf2
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: HYDROLASE Ligands: MG, ADP, BEF, MN |
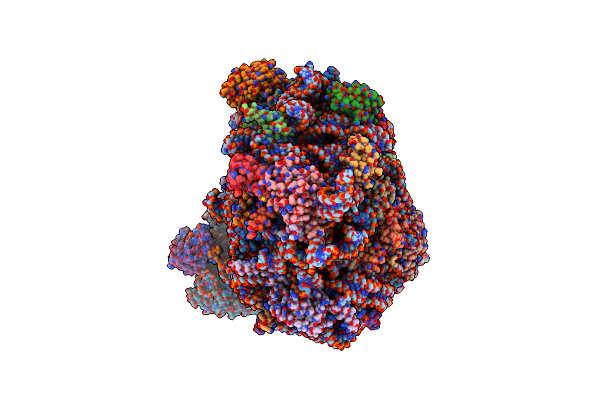 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: RIBOSOME Ligands: MG, K, NA, GTP, EPE, ZN |
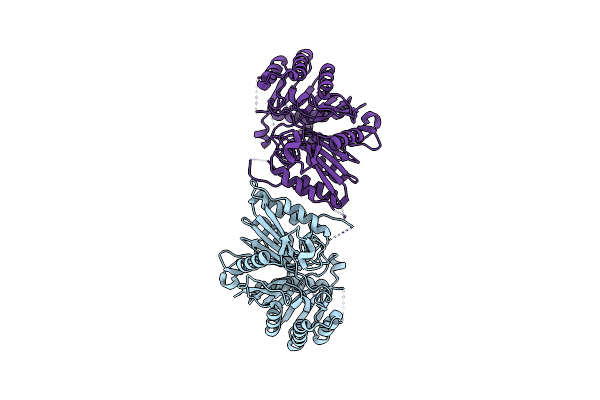 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of Glycerol Dehydrogenase In The Presence Of Nad+ And Glycerol
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-03-27 Classification: OXIDOREDUCTASE Ligands: ZN, NAD, GOL |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2023-06-14 Classification: OXIDOREDUCTASE Ligands: ZN, TRS |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-06-14 Classification: OXIDOREDUCTASE Ligands: ZN, NAD, TRS |
 |
Chaetomium Thermophilum Mre11-Rad50-Nbs1 Complex Bound To Atpys (Composite Structure)
Organism: Thermochaetoides thermophila
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: HYDROLASE Ligands: MN, MG, AGS |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: HYDROLASE Ligands: MN |
 |
Organism: Thermochaetoides thermophila
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2022-12-28 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2021-03-24 Classification: PROTEIN TRANSPORT Ligands: ZN, GOL |
 |
Organism: Aequorea victoria
Method: ELECTRON MICROSCOPY Release Date: 2019-01-23 Classification: LUMINESCENT PROTEIN |
 |
Organism: Thermobaculum terrenum
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-11-19 Classification: HYDROLASE Ligands: NI |
 |
Crystal Structure Of Crispr-Associated Protein In Complex With 2'-Deoxyadenosine 5'-Triphosphate
Organism: Thermobaculum terrenum
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2014-11-19 Classification: HYDROLASE Ligands: NI, MG, DTP |
 |
Crystal Structures Of Yrda From Escherichia Coli, A Homologous Protein Of Gamma-Class Carbonic Anhydrase, Show Possible Allosteric Conformations
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2012-08-01 Classification: TRANSFERASE Ligands: ZN, PO4 |
 |
Crystal Structures Of Yrda From Escherichia Coli, A Homologous Protein Of Gamma-Class Carbonic Anhydrases, Show Possible Allosteric Conformations
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-08-01 Classification: TRANSFERASE Ligands: ZN |
 |
Molecular Insights Into Plant Cell Proliferation Disturbance By Agrobacterium Protein 6B
Organism: Agrobacterium vitis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2011-01-26 Classification: TOXIN |

