Search Count: 18
 |
Organism: Arabidopsis thaliana, Aequorea victoria
Method: ELECTRON MICROSCOPY Release Date: 2023-11-22 Classification: MEMBRANE PROTEIN Ligands: CL, Y01 |
 |
Organism: Arabidopsis thaliana, Aequorea victoria
Method: ELECTRON MICROSCOPY Release Date: 2023-11-22 Classification: MEMBRANE PROTEIN Ligands: Y01, CL |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2023-11-15 Classification: MEMBRANE PROTEIN Ligands: CL, Y01 |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2023-11-15 Classification: MEMBRANE PROTEIN Ligands: CL, Y01 |
 |
Organism: Escherichia coli k-12, Rhodospirillum rubrum atcc 11170
Method: ELECTRON MICROSCOPY Release Date: 2022-01-12 Classification: CHAPERONE |
 |
X-Ray Crystal Structure Of Pyrococcus Furiosus General Transcription Factor Tfe-Alpha
Organism: Pyrococcus furiosus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-07-08 Classification: TRANSCRIPTION |
 |
X-Ray Crystal Structure Of Pyrococcus Furiosus General Transcription Factor Tfe-Alpha (Semet Labeled Protein)
Organism: Pyrococcus furiosus (strain atcc 43587 / dsm 3638 / jcm 8422 / vc1)
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2020-07-08 Classification: TRANSCRIPTION |
 |
Organism: Thermococcus kodakarensis (strain atcc baa-918 / jcm 12380 / kod1)
Method: ELECTRON MICROSCOPY Release Date: 2020-07-01 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
Organism: Thermococcus kodakarensis (strain atcc baa-918 / jcm 12380 / kod1)
Method: ELECTRON MICROSCOPY Release Date: 2020-07-01 Classification: TRANSCRIPTION Ligands: MG, ZN |
 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2017-12-27 Classification: TRANSFERASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2017-12-27 Classification: TRANSFERASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2017-12-27 Classification: TRANSFERASE Ligands: EDO, MN, UDP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.64 Å Release Date: 2017-12-20 Classification: TRANSFERASE Ligands: UDP, MN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2017-12-20 Classification: TRANSFERASE Ligands: UD1, MN |
 |
Organism: Nonlabens marinus s1-08
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-11-23 Classification: TRANSPORT PROTEIN Ligands: ANR, CL, OLA |
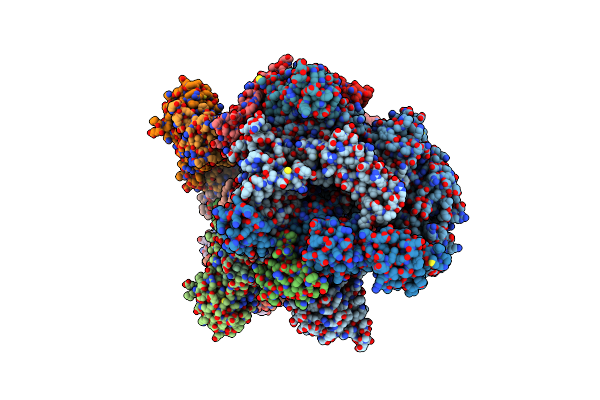 |
Crystal Structure Of Euryarchaeal Rna Polymerase From Thermococcus Kodakarensis
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2014-10-08 Classification: TRANSCRIPTION Ligands: MG, ZN |
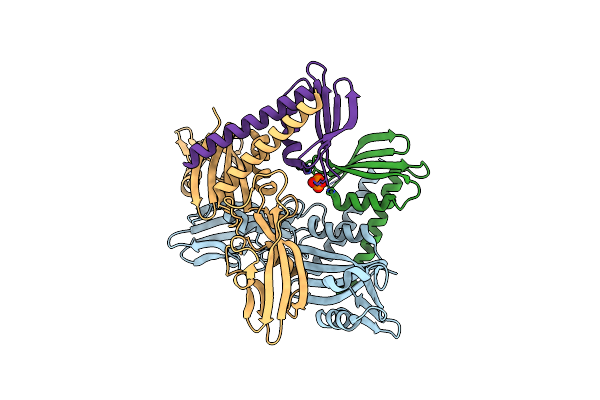 |
The X-Ray Crystal Structure Of Rpo3/Rpo11 Heterodimer Of Euryarchaeal Rna Polymerase From Thermococcus Kodakarensis
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-10-08 Classification: TRANSFERASE Ligands: PO4 |

