Search Count: 21
 |
Organism: Aeromonas hydrophila
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: TOXIN |
 |
Organism: Aeromonas hydrophila
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: TOXIN |
 |
Organism: Aeromonas hydrophila
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: TOXIN |
 |
Organism: Aeromonas hydrophila
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: TOXIN |
 |
Organism: Aeromonas hydrophila
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: TOXIN |
 |
Organism: Drosophila melanogaster
Method: SOLUTION NMR Release Date: 2024-04-17 Classification: UNKNOWN FUNCTION |
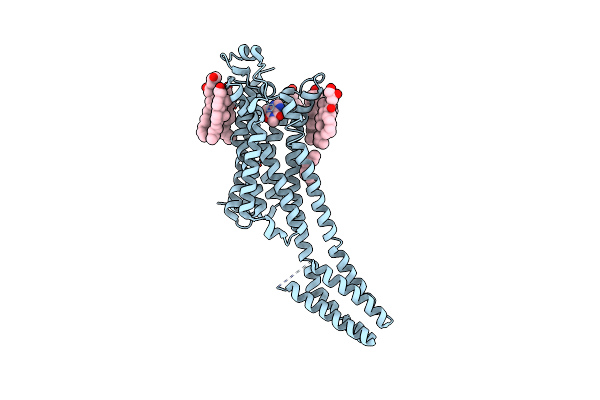 |
Room-Temperature Structure Of The Stabilised A2A-Theophylline Complex Determined By Synchrotron Serial Crystallography
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2023-08-30 Classification: MEMBRANE PROTEIN Ligands: TEP, CLR, OLA, OLC, NA |
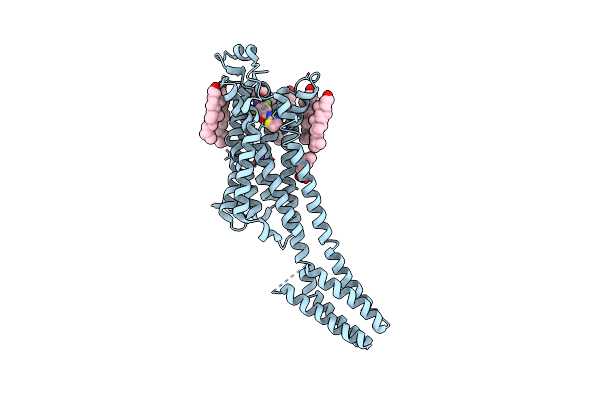 |
Room-Temperature Structure Of The Stabilised A2A-Luaa47070 Complex Determined By Synchrotron Serial Crystallography
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2023-08-30 Classification: MEMBRANE PROTEIN Ligands: 9Y2, CLR, OLA, NA |
 |
Room Temperature Structure Of Archaerhodopsin-3 Obtained 110 Ns After Photoexcitation
Organism: Halorubrum sodomense
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-06-14 Classification: PROTON TRANSPORT Ligands: RET, DD9, R16, PLM, CA, NA, MG, CL |
 |
Dark-Adapted Structure Of Archaerhodopsin-3 Obtained From Lcp Crystals Using A Thin-Film Sandwich At Room Temperature
Organism: Halorubrum sodomense
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-07-22 Classification: PROTON TRANSPORT Ligands: RET, R16, DD9, CA, NA, CL |
 |
Organism: Halorubrum sodomense
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2020-07-22 Classification: PROTON TRANSPORT Ligands: RET, R16, D12, DD9, CA, NA, CL |
 |
Organism: Halorubrum sodomense
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2019-10-09 Classification: PROTON TRANSPORT Ligands: RET, R16, D12, DD9, CA, NA, CL |
 |
Room Temperature Structure Of Archaerhodopsin-3 Via Lcp Extruder Using Synchrotron Radiation
Organism: Halorubrum sodomense
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-10-09 Classification: PROTON TRANSPORT Ligands: RET, R16, DD9, CA, NA, CL |
 |
Ground State Structure Of Archaerhodopsin-3 Obtained From Lcp Crystals Using A Thin-Film Sandwich At Room Temperature
Organism: Halorubrum sodomense
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-10-09 Classification: PROTON TRANSPORT Ligands: RET, R16, DD9, D10, CA, NA, CL |
 |
Egfr Kinase Domain In Complex With A Covalent Aminobenzimidazole Inhibitor.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2016-07-27 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 5X4 |
 |
Egfr Kinase Domain T790M Mutant In Complex With A Covalent Aminobenzimidazole Inhibitor.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2016-07-27 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 5X4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2016-07-27 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 5XH |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2016-06-08 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: MG, ANP, 57N |
 |
Organism: Azoarcus sp. cib
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2014-12-24 Classification: TRANSCRIPTION |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2014-02-26 Classification: IMMUNE SYSTEM |

