Search Count: 26
 |
Organism: Rhodobacteraceae bacterium qy30, Dna molecule
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: ANTIVIRAL PROTEIN/DNA Ligands: MN |
 |
Organism: Rhodobacteraceae bacterium qy30
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: ANTIVIRAL PROTEIN/DNA Ligands: PO4 |
 |
Organism: Rhodobacteraceae bacterium qy30
Method: X-RAY DIFFRACTION Release Date: 2025-05-07 Classification: ANTIVIRAL PROTEIN Ligands: A1A3G, F2A, MN, A1A3H, D5M |
 |
Organism: Rhodobacteraceae bacterium qy30, Dna molecule
Method: ELECTRON MICROSCOPY Release Date: 2025-05-07 Classification: ANTIVIRAL PROTEIN |
 |
Organism: Cupriavidus metallidurans ch34
Method: ELECTRON MICROSCOPY Release Date: 2024-05-08 Classification: ISOMERASE Ligands: GDP, MG |
 |
Organism: Cupriavidus metallidurans ch34
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: ISOMERASE |
 |
Organism: Rattus norvegicus, Gallus gallus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-10-11 Classification: CELL CYCLE Ligands: GTP, MG, CA, GOL, GDP, 6FS, MES, ACP |
 |
Structure Of The Catalytic Domain Of The Class I Polyhydroxybutyrate Synthase From Cupriavidus Necator
Organism: Cupriavidus necator
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2016-10-26 Classification: BIOSYNTHETIC PROTEIN Ligands: SO4 |
 |
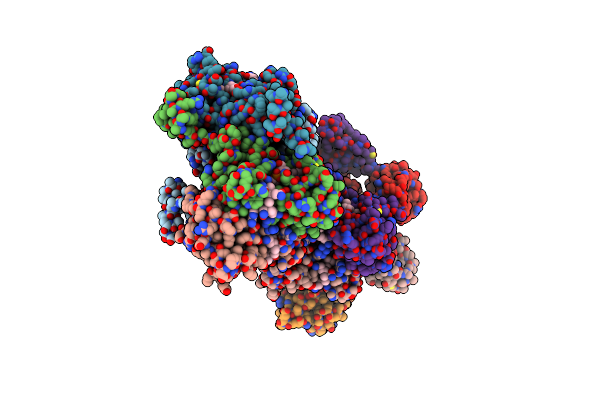
Crystal Structure Of A Truncated Form Of Thermus Thermophilus Carh Bound To Adenosylcobalamin (Dark State)
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2015-09-30 Classification: TRANSCRIPTIONAL REGULATOR Ligands: B12, 5AD, GOL |
 |
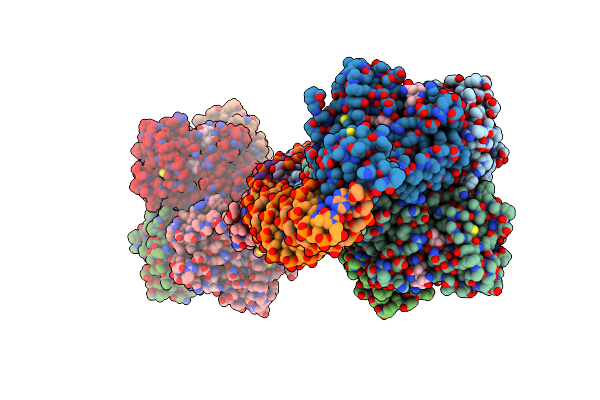
Crystal Structure Of Full-Length Thermus Thermophilus Carh Bound To Adenosylcobalamin (Dark State)
Organism: Thermus thermophilus (strain hb27 / atcc baa-163 / dsm 7039)
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2015-09-30 Classification: TRANSCRIPTIONAL REGULATOR Ligands: B12, 5AD |
 |
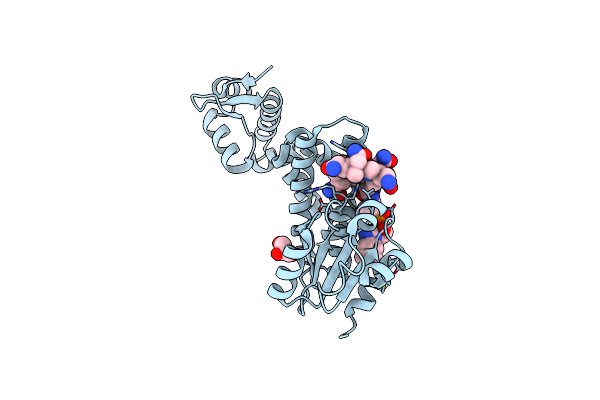
Crystal Structure Of Thermus Thermophilus Carh Bound To Adenosylcobalamin And A 26-Bp Dna Segment
Organism: Thermus thermophilus (strain hb27 / atcc baa-163 / dsm 7039), Thermus thermophilus hb27
Method: X-RAY DIFFRACTION Resolution:3.89 Å Release Date: 2015-09-30 Classification: Transcription regulator/DNA Ligands: B12, 5AD |
 |
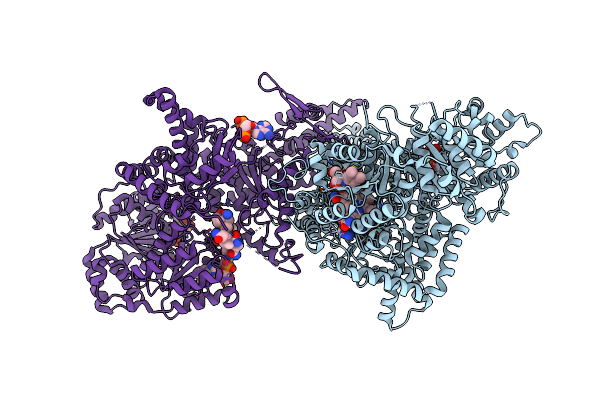
Crystal Structure Of Light-Exposed Full-Length Thermus Thermophilus Carh Bound To Cobalamin
Organism: Thermus thermophilus (strain hb27 / atcc baa-163 / dsm 7039)
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2015-09-30 Classification: TRANSCRIPTIONAL REGULATOR Ligands: B12, CL, GOL |
 |
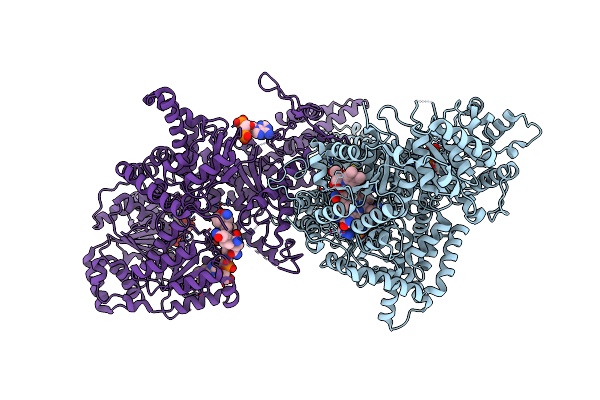
Isobutyryl-Coa Mutase Fused With Bound Adenosylcobalamin, Gdp, Mg (Holo-Icmf/Gdp), And Substrate Isobutyryl-Coenzyme A
Organism: Ralstonia metallidurans (strain ch34 / atcc 43123 / dsm 2839)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2015-09-09 Classification: ISOMERASE Ligands: B12, 5AD, CO6, GDP, MG |
 |
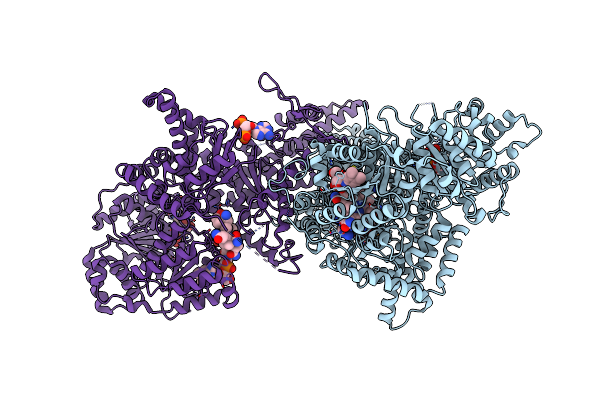
Isobutyryl-Coa Mutase Fused With Bound Adenosylcobalamin, Gdp, Mg (Holo-Icmf/Gdp), And Substrate N-Butyryl-Coenzyme A
Organism: Ralstonia metallidurans (strain ch34 / atcc 43123 / dsm 2839)
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2015-09-09 Classification: ISOMERASE Ligands: B12, 5AD, BCO, GDP, MG |
 |
Isobutyryl-Coa Mutase Fused With Bound Adenosylcobalamin, Gdp, Mg (Holo-Icmf/Gdp), And Substrate Isovaleryl-Coenzyme A
Organism: Ralstonia metallidurans (strain ch34 / atcc 43123 / dsm 2839)
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2015-09-09 Classification: ISOMERASE Ligands: B12, 5AD, IVC, GDP, MG |
 |
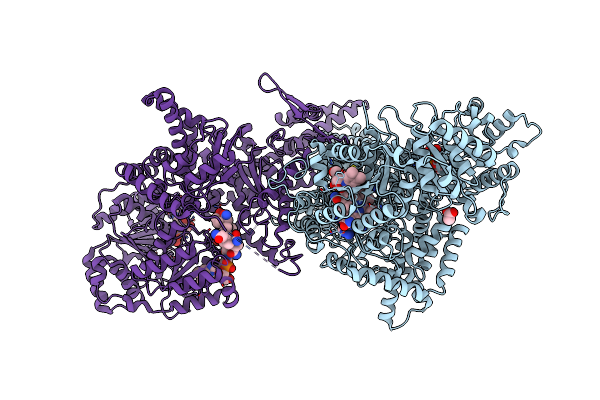
Isobutyryl-Coa Mutase Fused With Bound Adenosylcobalamin, Gdp, Mg (Holo-Icmf/Gdp), And Substrate Pivalyl-Coenzyme A
Organism: Ralstonia metallidurans (strain ch34 / atcc 43123 / dsm 2839)
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2015-09-09 Classification: ISOMERASE Ligands: B12, 5AD, 52O, GDP, MG |
 |
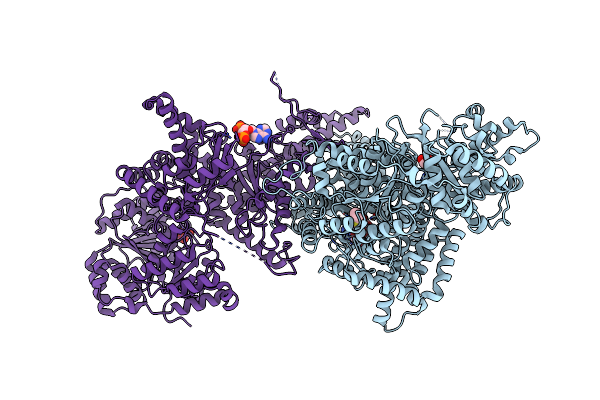
Isobutyryl-Coa Mutase Fused With Bound Adenosylcobalamin, Gdp, And Mg (Holo-Icmf/Gdp)
Organism: Ralstonia metallidurans (strain ch34 / atcc 43123 / dsm 2839)
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2015-02-11 Classification: ISOMERASE Ligands: B12, 5AD, GDP, MG, ACT |
 |
Isobutyryl-Coa Mutase Fused With Bound Butyryl-Coa And Without Cobalamin Or Gdp (Apo-Icmf)
Organism: Ralstonia metallidurans (strain ch34 / atcc 43123 / dsm 2839)
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2015-02-11 Classification: ISOMERASE Ligands: BCO, TLA |
 |
Isobutyryl-Coa Mutase Fused With Bound Butyryl-Coa, Gdp, And Mg And Without Cobalamin (Apo-Icmf/Gdp)
Organism: Ralstonia metallidurans (strain ch34 / atcc 43123 / dsm 2839)
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2015-02-11 Classification: ISOMERASE Ligands: BCO, GDP, MG |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.57 Å Release Date: 2013-02-27 Classification: HYDROLASE Ligands: ZN, SO4 |

