Search Count: 31
 |
Organism: Drosophila melanogaster, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2024-07-03 Classification: METAL BINDING PROTEIN Ligands: DMS, ZN, SO4 |
 |
Organism: Camelidae, Simian virus 12
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2023-11-22 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Resolution:9.60 Å Release Date: 2020-03-04 Classification: MEMBRANE PROTEIN |
 |
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2020-03-04 Classification: MEMBRANE PROTEIN |
 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719)
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2020-01-22 Classification: PEPTIDE BINDING PROTEIN Ligands: ZN |
 |
Organism: Arabidopsis thaliana, Glycine max
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2019-03-27 Classification: NUCLEAR PROTEIN |
 |
Crystal Structure Of Human Ash2L (Spry Domain And Sdi Motif) In Complex With Full Length Dpy-30
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2018-08-08 Classification: PROTEIN BINDING |
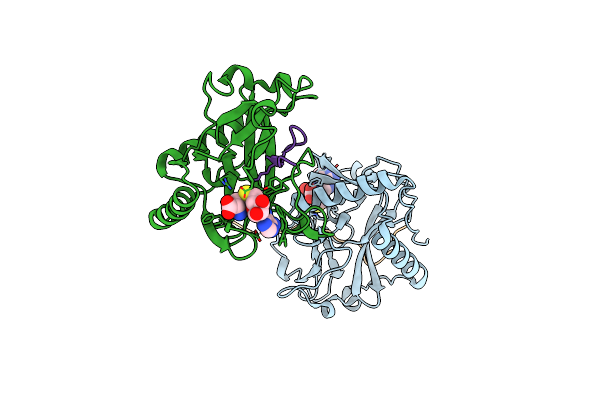 |
Crystal Structure Of Atxr5 In Complex With Histone H3.1 Mono-Methylated On R26
Organism: Ricinus communis, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-04-19 Classification: TRANSFERASE/DNA BINDING PROTEIN Ligands: SAH |
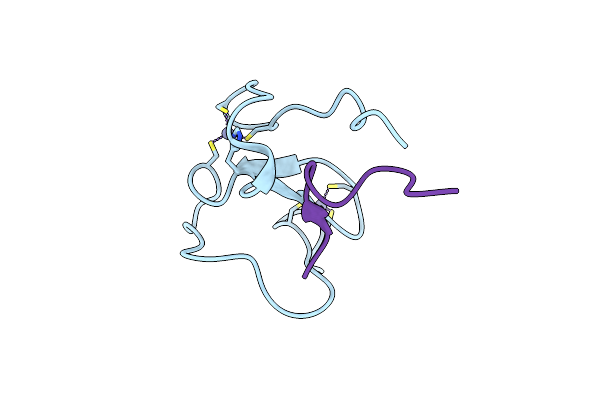 |
Organism: Glycine max, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-04-19 Classification: TRANSFERASE/DNA BINDING PROTEIN Ligands: ZN |
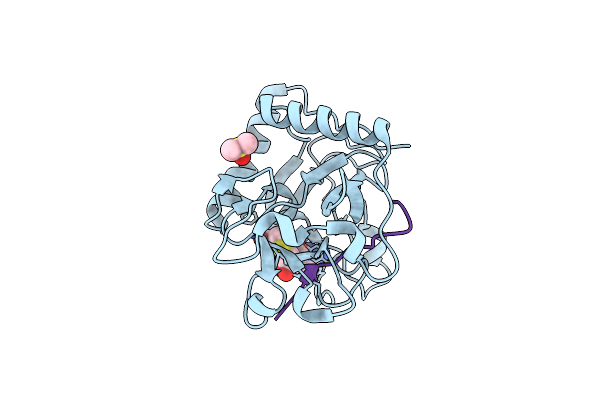 |
Crystal Structure Of Atxr5 Set Domain In Complex With K36Me3 Histone H3 Peptide
Organism: Ricinus communis, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2017-04-19 Classification: TRANSFERASE/DNA BINDING PROTEIN Ligands: SAH, DMS |
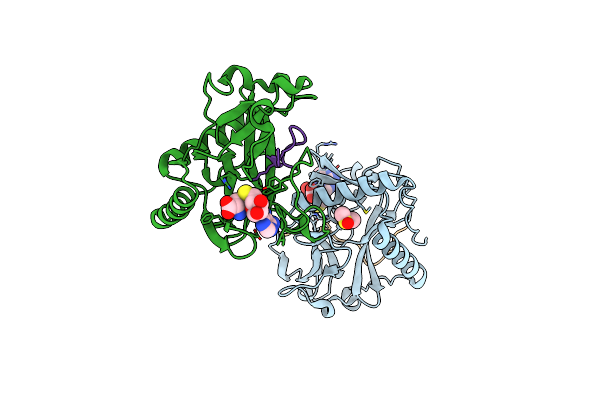 |
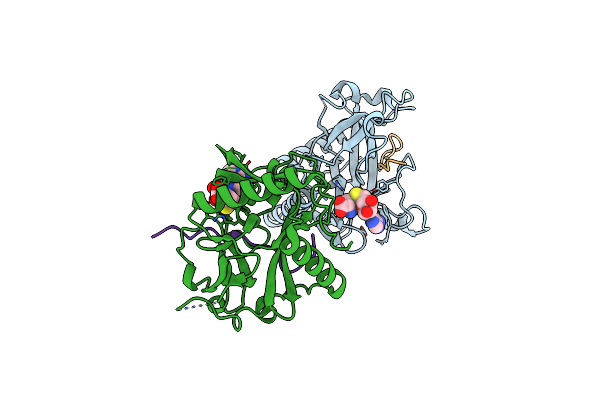
Organism: Ricinus communis, Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-04-19 Classification: TRANSFERASE/DNA BINDING PROTEIN Ligands: SAH, DMS |
 |
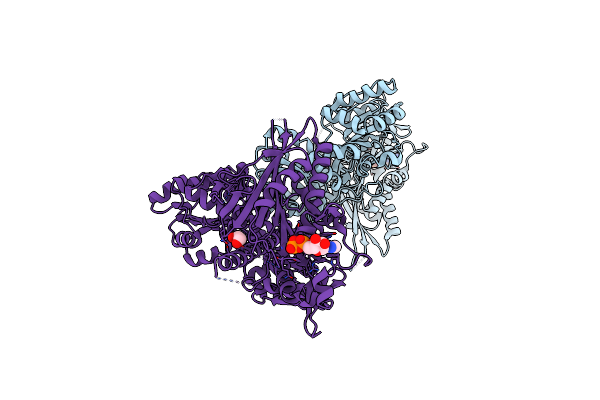
Crystal Structure Of Atxr5 Set Domain In Complex With Histone H3 Di-Methylated On R26
Organism: Ricinus communis, Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-04-05 Classification: TRANSFERASE/DNA BINDING PROTEIN Ligands: SAH |
 |
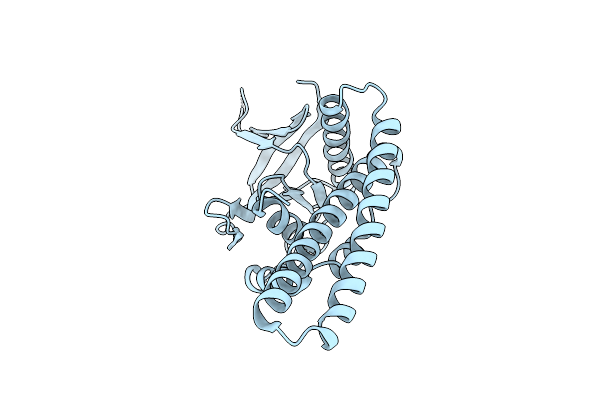
Organism: Escherichia coli umea 3304-1, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2014-07-09 Classification: MOTOR PROTEIN Ligands: ADP, EDO |
 |
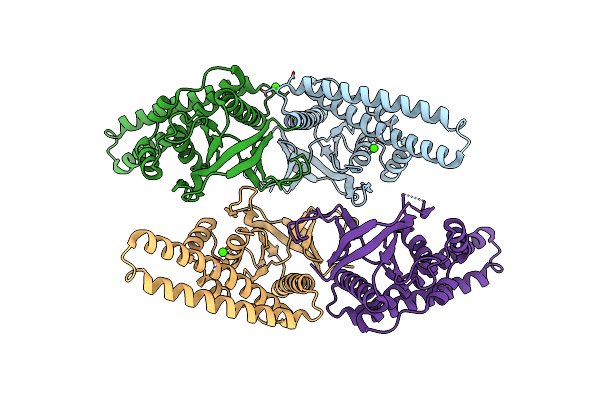
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2013-07-10 Classification: SIGNALING PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2013-07-10 Classification: SIGNALING PROTEIN Ligands: CA |
 |
Structure Of Candida Albicans Kar3 Motor Domain Fused To Maltose-Binding Protein
Organism: Escherichia coli, Candida albicans
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2012-10-10 Classification: MOTOR PROTEIN Ligands: MG, ADP, EDO |
 |
Structure Of The Truncated Neck And C-Terminal Motor Homology Domain Of Vik1 From Candida Glabrata
Organism: Candida glabrata
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2012-10-03 Classification: STRUCTURAL PROTEIN |
 |
Structure Of The Neck And C-Terminal Motor Homology Domain Of Vik1 From Candida Glabrata
Organism: Candida glabrata
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2012-10-03 Classification: STRUCTURAL PROTEIN Ligands: HOH |
 |
Organism: Candida glabrata
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2012-10-03 Classification: STRUCTURAL PROTEIN Ligands: MG, ADP, EDO |
 |
Organism: Drosophila melanogaster
Method: SOLUTION NMR Release Date: 2012-08-22 Classification: METAL BINDING PROTEIN Ligands: CA |

