Search Count: 33
 |
Organism: Rattus norvegicus, Mus musculus
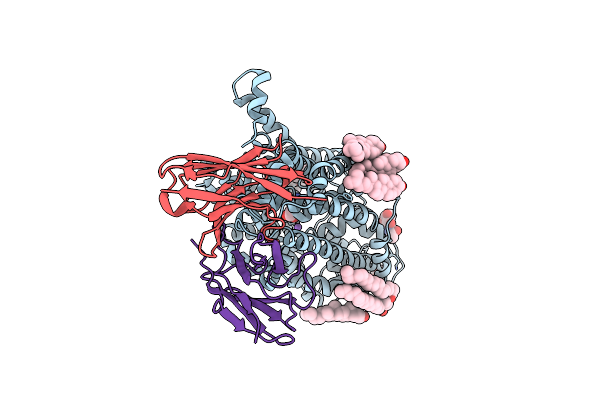
Method: ELECTRON MICROSCOPY Release Date: 2023-05-31 Classification: MEMBRANE PROTEIN Ligands: NAG, CL, NA, ABU, CLR, PTY |
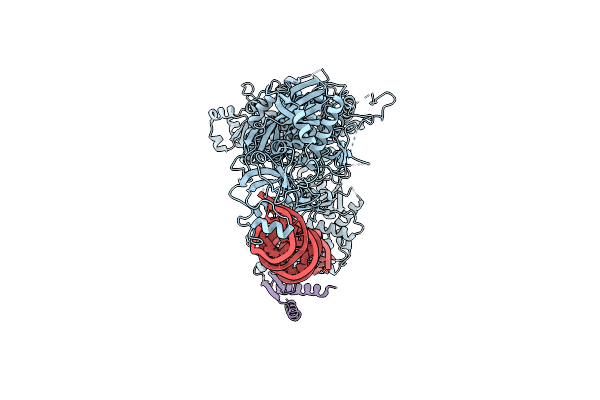 |
Mammalian Dicer In The "Pre-Dicing State" With Pre-Mir-15A Substrate And Tarbp2 Subunit
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-12-28 Classification: RNA BINDING PROTEIN |
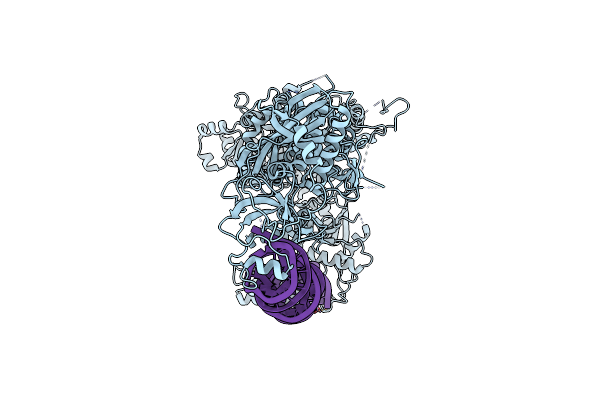 |
Organism: Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: RNA BINDING PROTEIN |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: RNA BINDING PROTEIN |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: RNA BINDING PROTEIN |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: RNA BINDING PROTEIN |
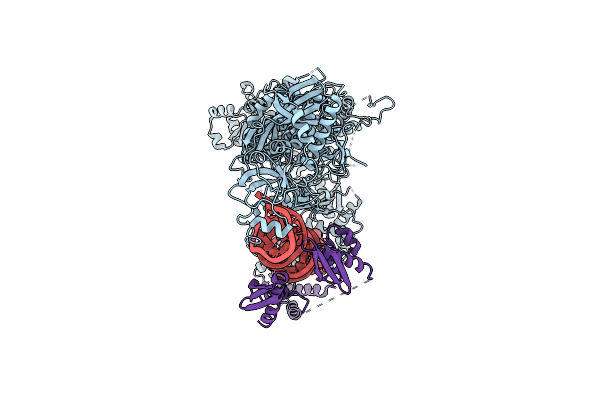 |
Mammalian Dicer In The "Pre-Dicing State" With Pre-Mir-15A Substrate And Tarbp2 Subunit
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-16 Classification: RNA BINDING PROTEIN |
 |
X-Ray Structure Of Thermostabilized Drosophila Dopamine Transporter With Gaba Transporter1-Like Substitutions In The Binding Site, In Substrate-Free Form.
Organism: Drosophila melanogaster, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2022-06-29 Classification: MEMBRANE PROTEIN |
 |
X-Ray Structure Of Thermostabilized Drosophila Dopamine Transporter With Gaba Transporter1-Like Substitutions In The Binding Site, In Complex With No711.
Organism: Drosophila melanogaster, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2022-06-29 Classification: MEMBRANE PROTEIN Ligands: 9BC, CLR, NA, CL, Y01 |
 |
X-Ray Structure Of Thermostabilized Drosophila Dopamine Transporter With Gaba Transporter1-Like Substitutions In The Binding Site, In Complex With Skf89976A
Organism: Drosophila melanogaster, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2022-06-29 Classification: MEMBRANE PROTEIN Ligands: CL, NA, 1WR, Y01, DMU, CLR |
 |
X-Ray Structure Of Drosophila Dopamine Transporter With Subsiteb Mutations (D121G/S426M) In Substrate-Free Form
Organism: Drosophila melanogaster, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2021-02-17 Classification: MEMBRANE PROTEIN Ligands: CL, CLR, P4G, NA |
 |
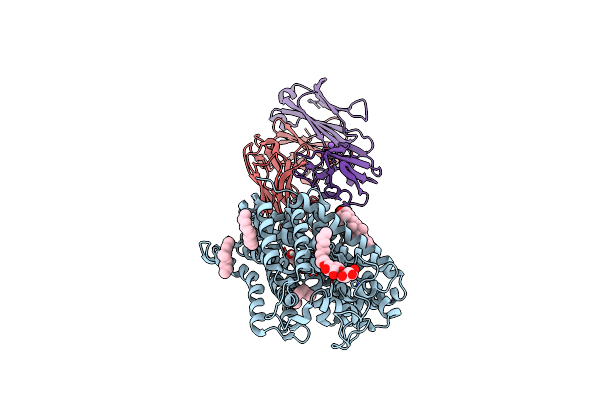
X-Ray Structure Of Drosophila Dopamine Transporter With Net-Like Mutations (D121G/S426M/F471L) In L-Norepinephrine Bound Form
Organism: Drosophila melanogaster, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.88 Å Release Date: 2021-02-17 Classification: MEMBRANE PROTEIN Ligands: CLR, Y01, DMU, NA, CL, LNR |
 |
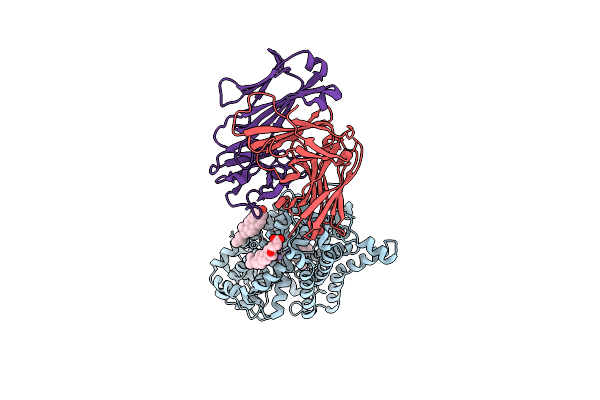
X-Ray Structure Of A Functional Drosophila Dopamine Transporter In L-Norepinephrine Bound Form
Organism: Drosophila melanogaster, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-02-17 Classification: MEMBRANE PROTEIN Ligands: DMU, Y01, CLR, LNR, CL, NA, D10 |
 |
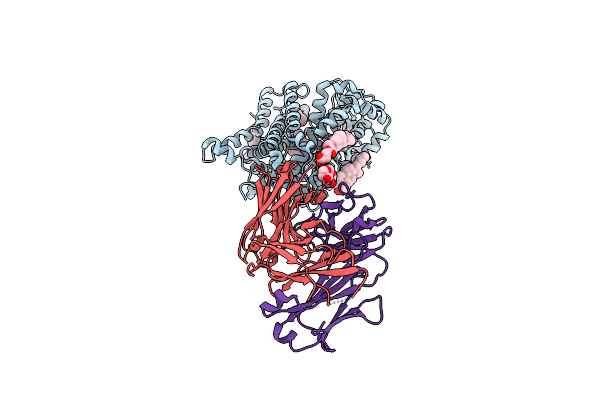
X-Ray Structure Of A Drosophila Dopamine Transporter With Subsiteb Mutations (D121G/S426M) In S-Duloxetine Bound Form
Organism: Drosophila melanogaster, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2021-02-17 Classification: MEMBRANE PROTEIN Ligands: 29E, Y01, NA, CL, DMU, CLR |
 |
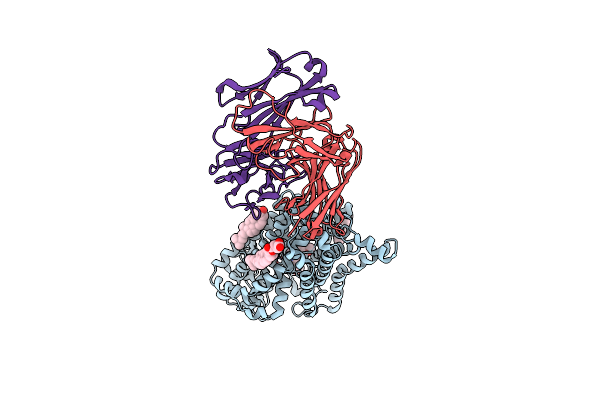
X-Ray Structure Of A Drosophila Dopamine Transporter With Net-Like Mutations (D121G/S426M/F471L) In Milnacipran Bound Form
Organism: Drosophila melanogaster, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2021-02-17 Classification: MEMBRANE PROTEIN Ligands: F0F, GOL, D10, CL, NA, Y01, CLR |
 |
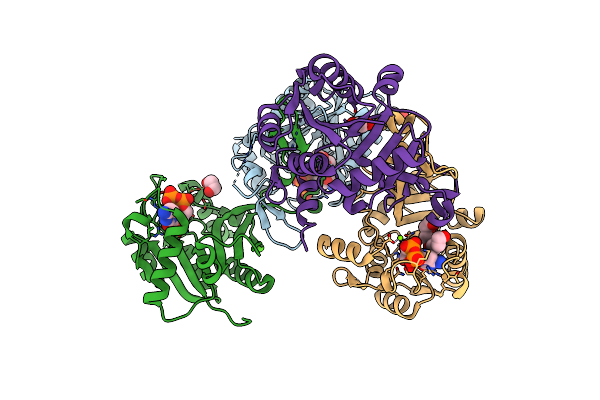
X-Ray Structure Of A Drosophila Dopamine Transporter With Net-Like Mutations (D121G/S426M/F471L) In Tramadol Bound Form
Organism: Drosophila melanogaster, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2021-02-17 Classification: MEMBRANE PROTEIN Ligands: CLR, Y01, NA, CL, F1U, D10 |
 |
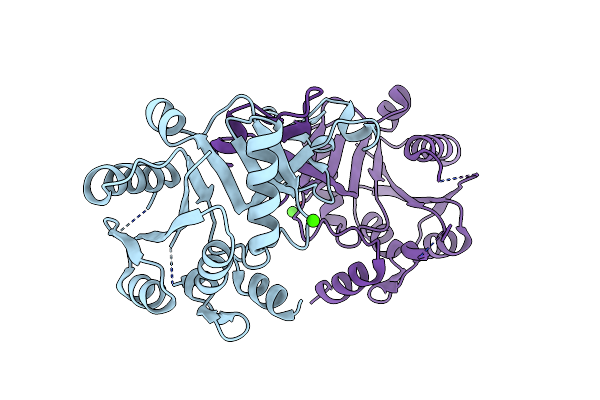
Crystal Structure Of Cmp-N-Acetylneuraminate Synthetase From Vibrio Cholerae In Complex With Cdp And Mg2+.
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-07-03 Classification: SUGAR BINDING PROTEIN Ligands: CDP, PGE, EDO, CA, MG, PG4 |
 |
Crystal Structure Of The Apo Form Of Cmp-N-Acetylneuraminate Synthetase From Vibrio Cholerae
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2019-07-03 Classification: SUGAR BINDING PROTEIN Ligands: CA |
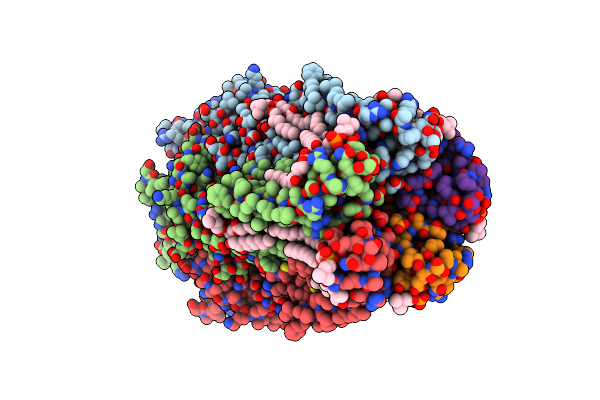 |
Organism: Gloeobacter violaceus (strain pcc 7421)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2017-03-29 Classification: MEMBRANE PROTEIN Ligands: LMT, PC1, ACT, 6E3, BR, NA |
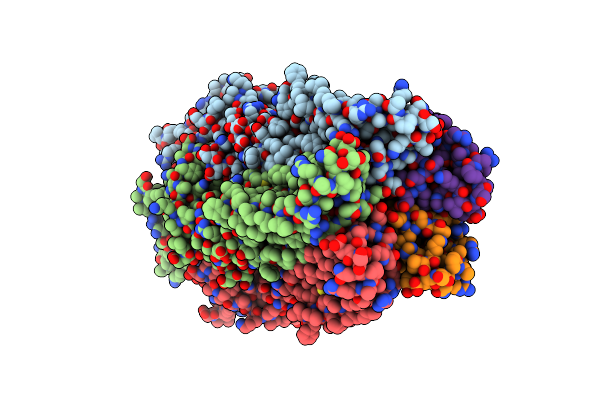 |
X-Ray Structure Of The 2-22' Locally-Closed Mutant Of Glic In Complex With Cyanoselenobarbital (Seleniated Barbiturate)
Organism: Gloeobacter
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2016-12-21 Classification: MEMBRANE PROTEIN, TRANSPORT PROTEIN Ligands: CL, ACT, NA, 6JA, D12 |

