Search Count: 27
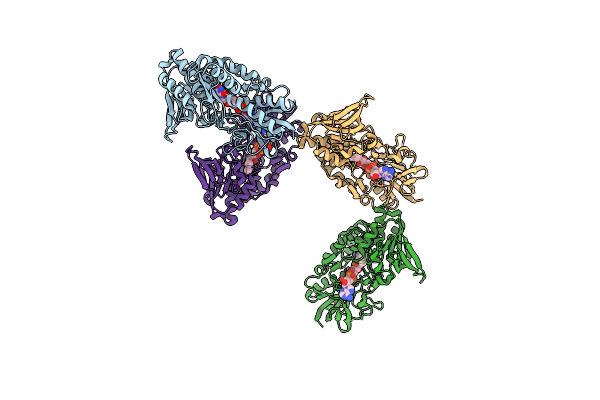 |
Crystal Structure Of The Selenomethionine-Substituted Human Sulfide:Quinone Oxidoreductase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2019-04-10 Classification: membrane protein, oxidoreductase Ligands: FAD, CL |
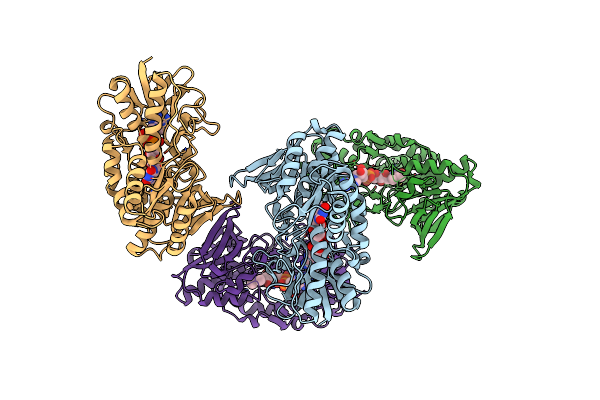 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.99 Å Release Date: 2019-04-10 Classification: membrane protein, oxidoreductase Ligands: FAD |
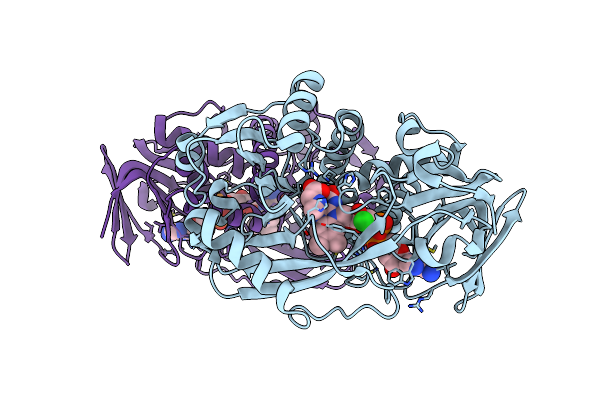 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2011-06-01 Classification: OXIDOREDUCTASE Ligands: FAD, CL, SAR |
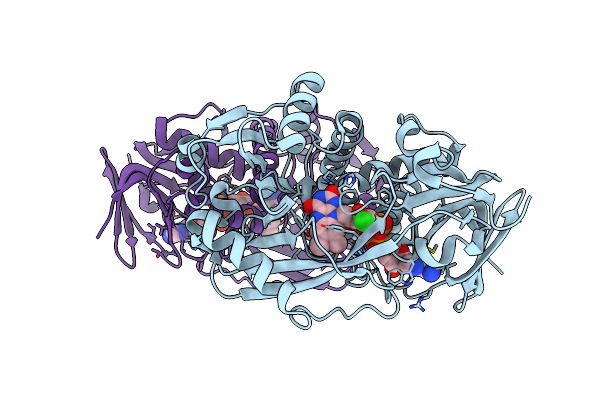 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-06-01 Classification: OXIDOREDUCTASE Ligands: FAD, CL |
 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2011-06-01 Classification: OXIDOREDUCTASE Ligands: FAD, CL, MTG |
 |
Crystal Structure Of Glucose Oxidase For Space Group C2221 At 1.2 A Resolution
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2011-06-01 Classification: OXIDOREDUCTASE Ligands: NAG, FAD, GOL, PEG, CL |
 |
Crystal Structure Of Glucose Oxidase For Space Group P3121 At 1.3 A Resolution.
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2011-06-01 Classification: OXIDOREDUCTASE Ligands: NAG, FAD, CL |
 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: FAD, CL |
 |
Crystal Structure Of The Lys265Arg Phosphate-Crytsallized Mutant Of Monomeric Sarcosine Oxidase
Organism: Bacillus sp. b-0618
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: FAD, CL |
 |
Crystal Structure Of The Lys265Arg Peg-Crystallized Mutant Of Monomeric Sarcosine Oxidase
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: FAD, CL, PO4 |
 |
Tyr258Phe Mutant Of Nikd, An Unusual Amino Acid Oxidase Essential For Nikkomycin Biosynthesis: Open Form At 1.55A Resolution
Organism: Streptomyces tendae
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2009-10-20 Classification: OXIDOREDUCTASE Ligands: FAD, 6PC, MPD |
 |
Nikd, An Unusual Amino Acid Oxidase Essential For Nikkomycin Biosynthesis: Closed Form At 1.15 A Resolution
Organism: Streptomyces tendae
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2007-07-31 Classification: OXIDOREDUCTASE |
 |
Nikd, An Unusual Amino Acid Oxidase Essential For Nikkomycin Biosynthesis: Open Form At 1.9A Resolution
Organism: Streptomyces tendae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2007-07-31 Classification: OXIDOREDUCTASE Ligands: FAD, 6PC, MPD |
 |
Organism: Streptomyces tendae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2007-07-31 Classification: FLAVOPROTEIN Ligands: FAD, BEZ |
 |
Structure Of The Complex Of Monomeric Sarcosine With Its Substrate Analogue Inhibitor 2-Furoic Acid At 1.3 A Resolution.
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2007-02-06 Classification: OXIDOREDUCTASE Ligands: CL, PO4, FAD, FOA, GOL |
 |
Heteroteterameric Sarcosine: Structure Of A Diflavin Metaloenzyme At 1.85 A Resolution
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2006-08-08 Classification: OXIDOREDUCTASE Ligands: FOA, NAD, GOL, SO3, FAD, FMN, ZN |
 |
Heterotetrameric Sarcosine: Structure Of A Diflavin Metaloenzyme At 1.85 A Resolution
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-08-08 Classification: OXIDOREDUCTASE Ligands: NAD, FAD, FMN, FOA, ZN |
 |
Monomeric Sarcosine Oxidase: Structure Of A Covalently Flavinylated Amine Oxidizing Enzyme
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2006-03-21 Classification: OXIDOREDUCTASE Ligands: PO4, CL, FAD |
 |
Monomeric Sarcosine Oxidase: Structure Of A Covalently Flavinylated Amine Oxidizing Enzyme
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2006-01-17 Classification: OXIDOREDUCTASE Ligands: CL, FCG, PO4 |
 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2002-08-30 Classification: OXIDOREDUCTASE Ligands: CL, FAD, PO4 |

