Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
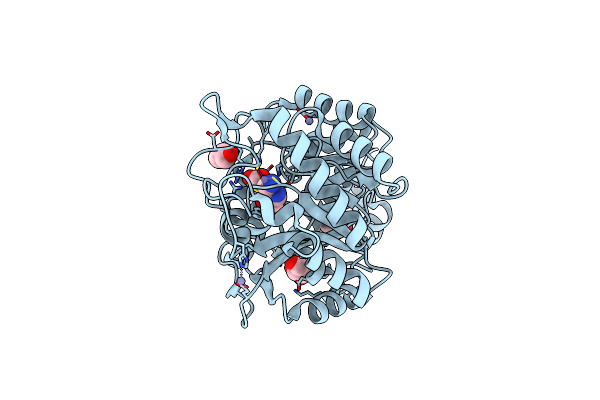 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-12-07 Classification: LYASE Ligands: PBG, ZN, GOL |
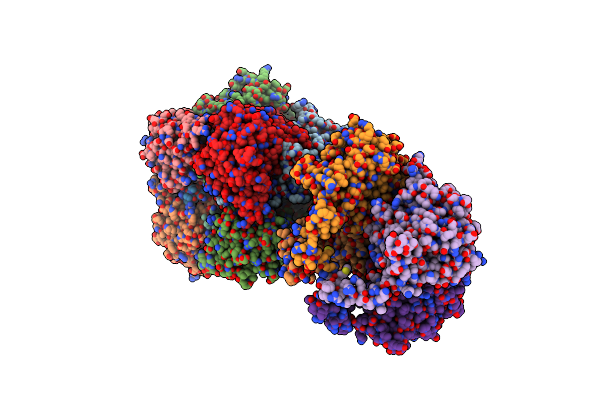 |
Organism: Pyrobaculum calidifontis (strain jcm 11548 / va1)
Method: X-RAY DIFFRACTION Resolution:3.47 Å Release Date: 2016-10-12 Classification: LYASE Ligands: ZN |
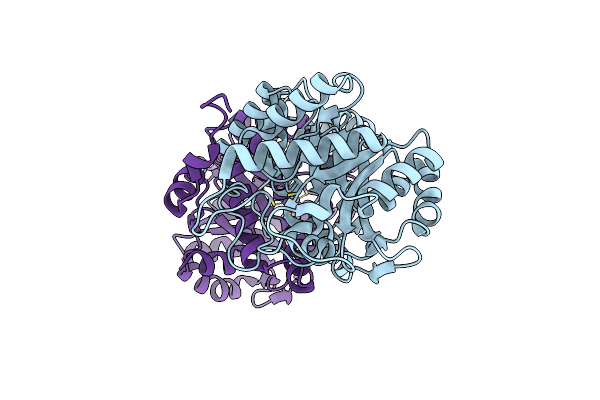 |
X-Ray Structure Of Human Recombinant 5-Aminolaevulinic Acid Dehydratase (Hralad).
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2016-01-27 Classification: LYASE Ligands: ZN |
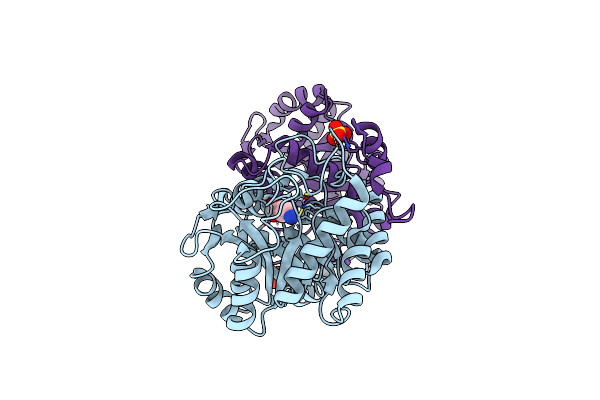 |
The X-Ray Structure Of Octameric Human Native 5-Aminolaevulinic Acid Dehydratase.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2016-01-27 Classification: LYASE Ligands: ZN, SO4, DAV |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2013-02-27 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 18W, ACT |
 |
Dynamic Domains Of Succinyl-Coa:3-Ketoacid-Coenzyme A Transferase From Pig Heart.
Organism: Sus scrofa
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2010-07-07 Classification: TRANSFERASE Ligands: CL, GOL |
 |
The Crystal Structure Of Human Porphobilinogen Deaminase At 2.8A Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2009-03-03 Classification: TRANSFERASE Ligands: DPM, SO4 |
 |
X-Ray Structure At 1.75 A Resolution Of A Norovirus Protease Linked To An Active Site Directed Peptide Inhibitor
Organism: Southampton virus (serotype 3)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2007-10-23 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: LGG |
 |
The X-Ray Structure Of Chlorobium Vibrioforme 5-Aminolaevulinic Acid Dehydratase Complexed With A Diacid Inhibitor
Organism: Chlorobium vibrioforme
Method: X-RAY DIFFRACTION Release Date: 2005-12-02 Classification: LYASE Ligands: MG, DSB |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2005-08-23 Classification: LYASE Ligands: SHO, ZN |
 |
Yeast 5-Aminolaevulinic Acid Dehydratase Putative Cyclic Reaction Intermediate Complex
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Release Date: 2003-06-02 Classification: LYASE Ligands: ZN, BME, PBG |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2001-08-02 Classification: DEHYDRATASE Ligands: DSB, ZN |
 |
Schiff-Base Complex Of Yeast 5-Aminolaevulinic Acid Dehydratase With 4-Oxosebacic Acid
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2001-08-02 Classification: DEHYDRATASE Ligands: 4OX, ZN |
 |
Schiff-Base Complex Of Yeast 5-Aminolaevulinic Acid Dehydratase With 4-Keto-5-Amino-Hexanoic (Kah) At 1.64 A Resolution
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2001-07-13 Classification: DEHYDRATASE Ligands: KAH, ZN |
 |
Crystal Structure Of Native Human Erythrocyte 5-Aminolaevulinic Acid Dehydratase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2001-07-12 Classification: LYASE Ligands: PBG, ZN, SO4 |
 |
Schiff-Base Complex Of Yeast 5-Aminolaevulinic Acid Dehydratase With Succinylacetone At 2.0 A Resolution.
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2001-07-12 Classification: DEHYDRATASE Ligands: SHU, ZN |
 |
Schiff-Base Complex Of Yeast 5-Aminolaevulinic Acid Dehydratase With Laevulinic Acid At 1.6 A Resolution
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2001-07-10 Classification: DEHYDRATASE Ligands: SHF, ZN |
 |
Schiff-Base Complex Of Yeast 5-Aminolaevulinic Acid Dehydratase With 5-Aminolaevulinic Acid At 1.7 A Resolution
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2001-07-10 Classification: DEHYDRATASE Ligands: DAV, ZN |
 |
Schiff-Base Complex Of Yeast 5-Aminolaevulinic Acid Dehydratase With Laevulinic Acid
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2000-02-23 Classification: LYASE Ligands: ZN, SHF |
 |
X-Ray Structure Of 5-Aminolevulinic Acid Dehydratase Complexed With The Inhibitor Levulinic Acid
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1999-12-17 Classification: LYASE Ligands: SO4, ZN, SHF, GOL |