Search Count: 34
 |
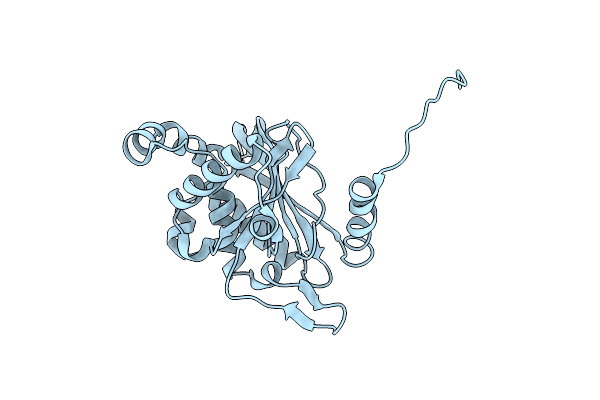
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-07-22 Classification: OXIDOREDUCTASE Ligands: TPP, MG |
 |
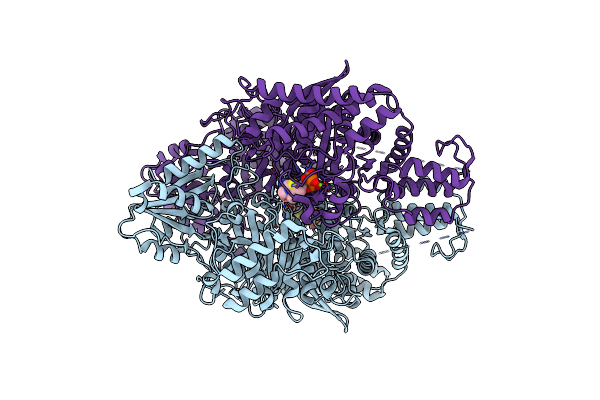
Cryo-Electron Microscopic Structure Of The Dihydrolipoamide Succinyltransferase (E2) Component Of The Human Alpha-Ketoglutarate (2-Oxoglutarate) Dehydrogenase Complex [Residues 218-453]
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-01-22 Classification: TRANSFERASE |
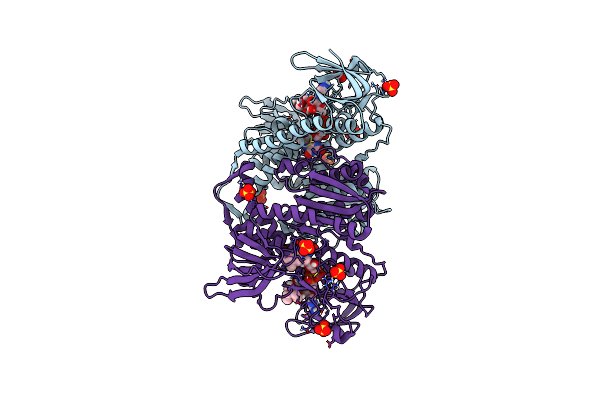 |
Crystal Structure Of The Disease-Causing G194C Mutant Of The Human Dihydrolipoamide Dehydrogenase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-11-20 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, BTB |
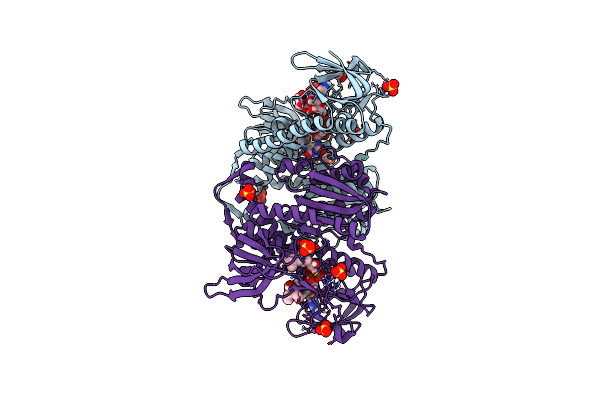 |
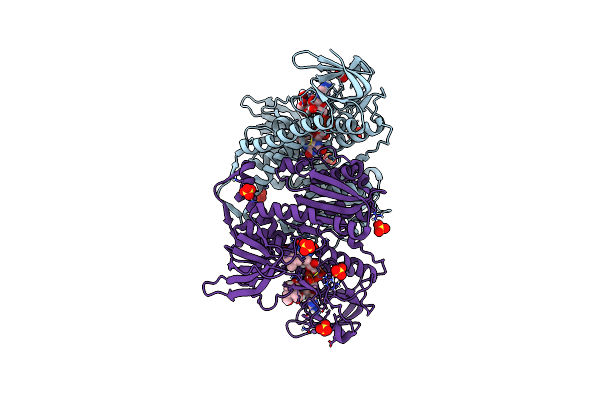
Crystal Structure Of The Human Dihydrolipoamide Dehydrogenase At 1.75 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-11-20 Classification: OXIDOREDUCTASE Ligands: FAD, BTB, SO4 |
 |
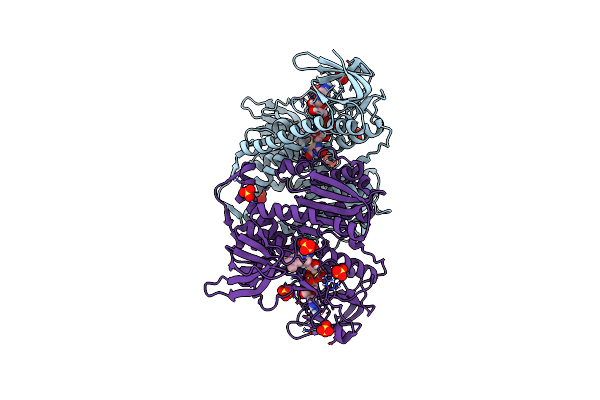
Crystal Structure Of The Disease-Causing R460G Mutant Of The Human Dihydrolipoamide Dehydrogenase At 1.44 Angstrom Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2019-11-20 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, BTB |
 |
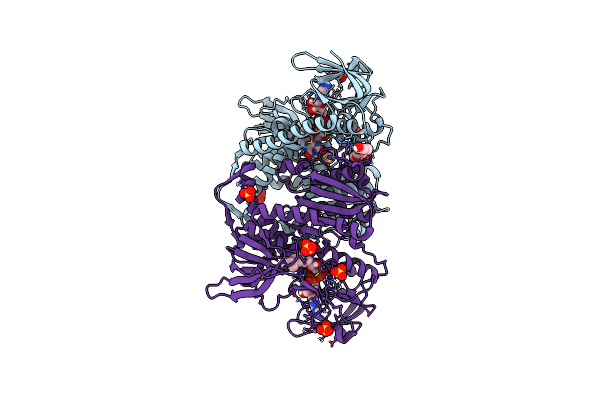
Crystal Structure Of The Disease-Causing R447G Mutant Of The Human Dihydrolipoamide Dehydrogenase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-11-20 Classification: OXIDOREDUCTASE Ligands: FAD, SO4 |
 |
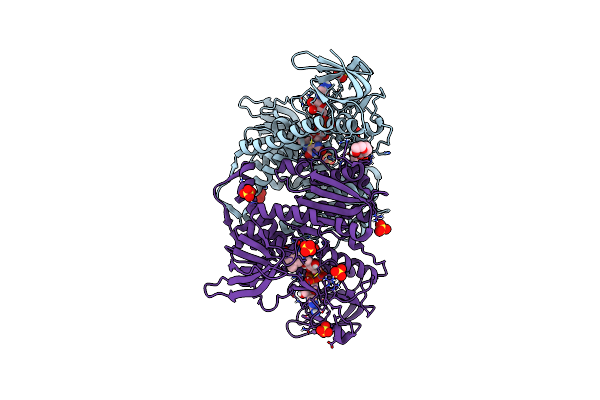
Crystal Structure Of The Disease-Causing I445M Mutant Of The Human Dihydrolipoamide Dehydrogenase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2019-11-20 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, 1PE |
 |
Crystal Structure Of The Disease-Causing G426E Mutant Of The Human Dihydrolipoamide Dehydrogenase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2019-11-20 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, 1PE |
 |
Crystal Structure Of The Disease-Causing P453L Mutant Of The Human Dihydrolipoamide Dehydrogenase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2019-11-20 Classification: OXIDOREDUCTASE Ligands: FAD, SO4 |
 |
Crystal Structure Of The R460G Disease-Causing Mutant Of The Human Dihydrolipoamide Dehydrogenase.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2019-09-04 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, TRS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2018-07-11 Classification: OXIDOREDUCTASE Ligands: TPP, MG |
 |
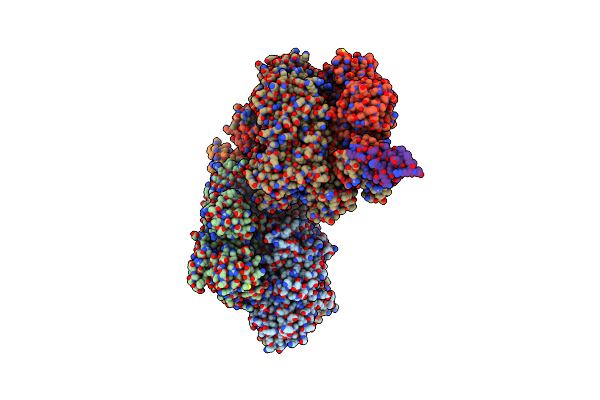
Human Pyruvate Dehydrogenase E1 Component Complex With Covalent Tdp Adduct Acetyl Phosphinate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2018-07-11 Classification: OXIDOREDUCTASE Ligands: A5X, MG |
 |
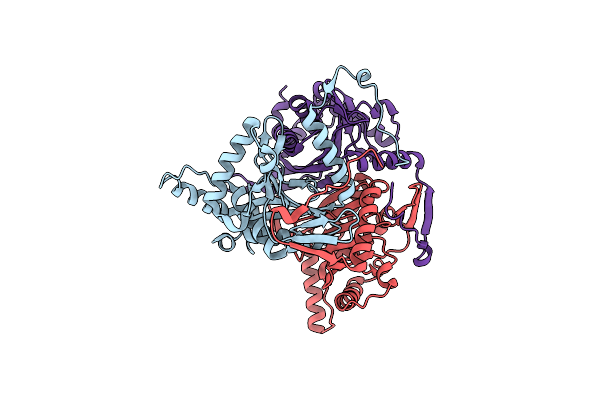
Novel Binding Motif And New Flexibility Revealed By Structural Analysis Of A Pyruvate Dehydrogenase-Dihydrolipoyl Acetyltransferase Sub-Complex From The Escherichia Coli Pyruvate Dehydrogenase Multi-Enzyme Complex
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-09-17 Classification: OXIDOREDUCTASE |
 |
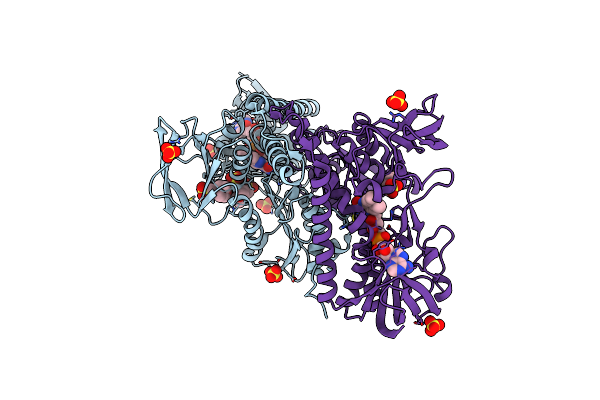
Catalytic Domain From Dihydrolipoamide Acetyltransferase Of Pyruvate Dehydrogenase From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2014-04-23 Classification: TRANSFERASE |
 |
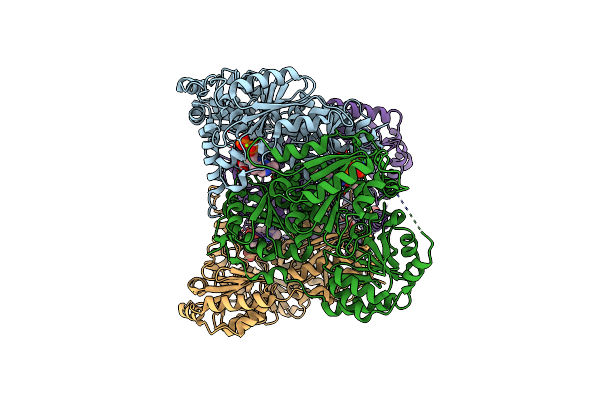
Dihydrolipoamide Dehydrogenase Of Pyruvate Dehydrogenase From Escherichia Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2013-04-24 Classification: OXIDOREDUCTASE Ligands: FAD, MES, SO4 |
 |
Pyruvate Decarboxylase Variant Glu473Asp From Z. Mobilis In Complex With Reaction Intermediate 2-Lactyl-Thdp
Organism: Zymomonas mobilis
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2010-09-08 Classification: LYASE Ligands: MG, TDL, GOL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-03-02 Classification: OXIDOREDUCTASE Ligands: EPE, TPP, MG, PO4 |
 |
E. Coli Pyruvate Dehydrogenase Complex E1 E235A Mutant With Low Tdp Concentration
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2010-03-02 Classification: OXIDOREDUCTASE Ligands: TPP, MG, PO4, EPE |
 |
E. Coli Pyruvate Dehydrogenase Complex E1 E235A Mutant With High Tdp Concentration
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2010-03-02 Classification: OXIDOREDUCTASE Ligands: TPP, MG, PO4, EPE |
 |
Crystal Structure Of Benzoylformate Decarboxylase In Complex With The Inhibitor Mbp
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2009-01-27 Classification: LYASE Ligands: D7K, CA |

