Search Count: 18
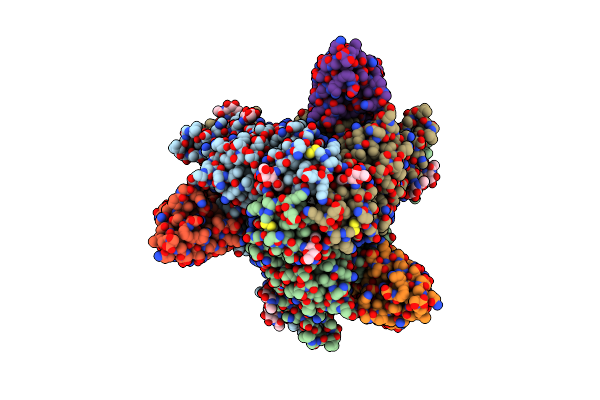 |
Prefusion F Glycoprotein Ectodomain Of Nipah Virus Ectodomain In Complex With Ds90 Nanobody
Organism: Henipavirus nipahense, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2025-06-04 Classification: VIRAL PROTEIN Ligands: NAG |
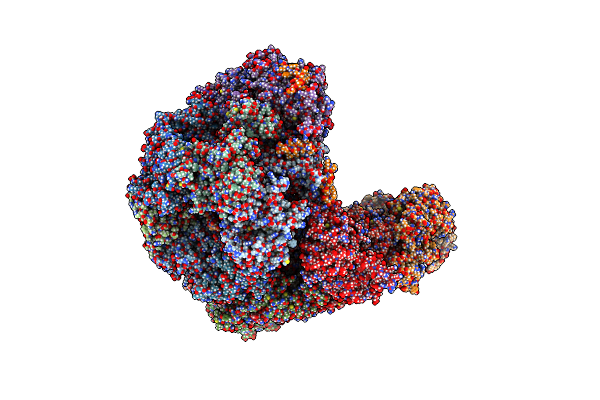 |
S. Cerevisiae Replisome + Ctf4, Bound By Pol Alpha Primase. Complex Engaged With A Fork Dna Substrate Containing A 60 Nucleotide Lagging Strand.
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: REPLICATION Ligands: ANP, MG, ZN |
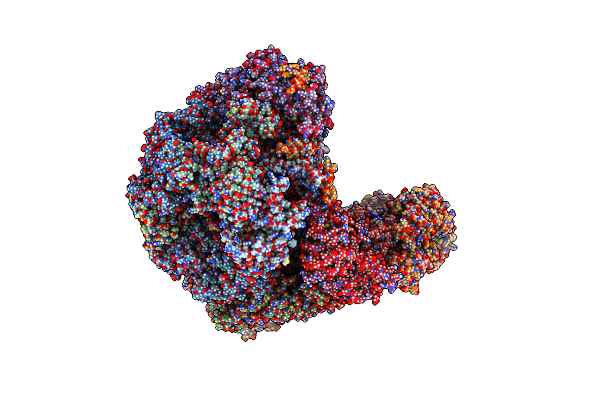 |
S. Cerevisiae Replisome + Ctf4, Bound By Pol Alpha. Complex Engaged With A Fork Dna Substrate Containing A 60 Nucleotide Lagging Strand.
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: REPLICATION Ligands: ANP, MG, ZN |
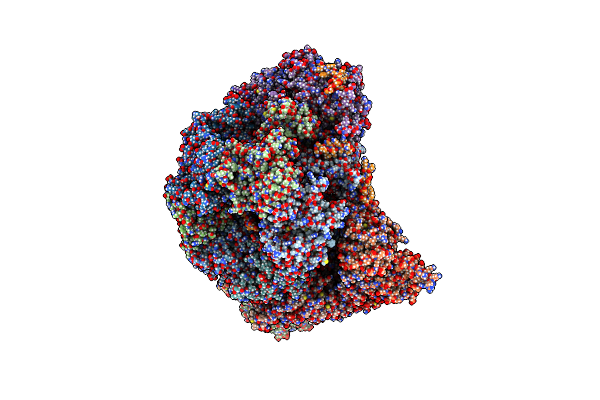 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: REPLICATION Ligands: ANP, MG, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-08-09 Classification: REPLICATION Ligands: ANP, MG, ZN |
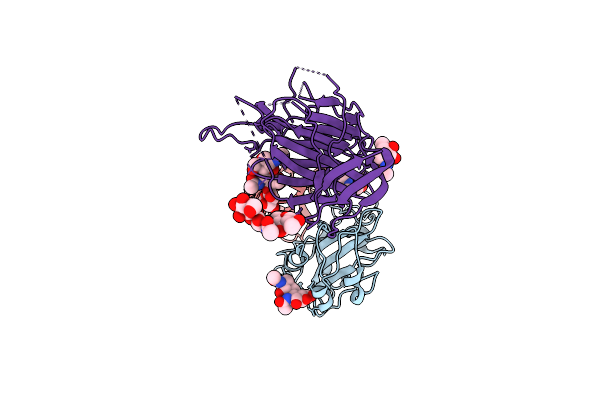 |
Cryo-Em Structure Of Sars-Cov-2 Spike (Hexapro Variant) In Complex With Nanobody W25 (Map 3, Focus Refinement On Rbd, W25 And Adjacent Ntd)
Organism: Severe acute respiratory syndrome coronavirus 2, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2023-03-08 Classification: VIRAL PROTEIN Ligands: NAG |
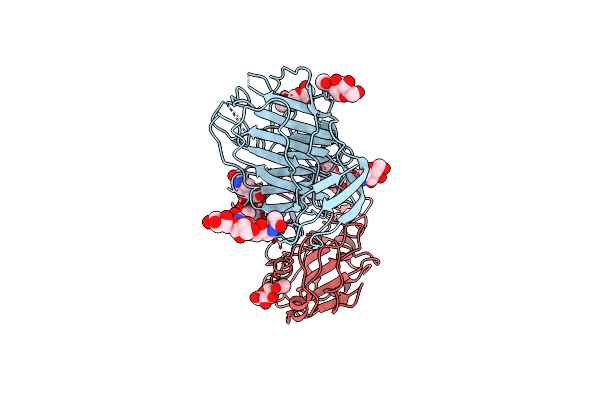 |
Cryo-Em Structure Of Sars-Cov-2 Spike (Omicron Ba.1 Variant) In Complex With Nanobody W25 (Map 5, Focus Refinement On Rbd, W25 And Adjacent Ntd)
Organism: Severe acute respiratory syndrome coronavirus 2, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2023-03-08 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-11-10 Classification: REPLICATION Ligands: ZN, MG, ANP, SO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-11-10 Classification: REPLICATION Ligands: ZN, MG, ANP, SO4 |
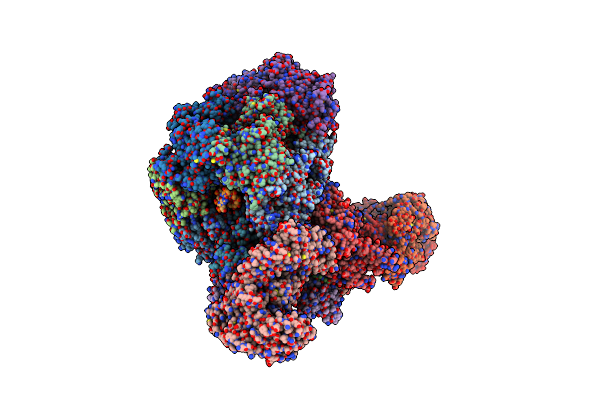 |
S. Cerevisiae Replisome-Scf(Dia2) Complex Bound To Double-Stranded Dna (Conformation I)
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-11-10 Classification: REPLICATION Ligands: ANP, MG, ZN |
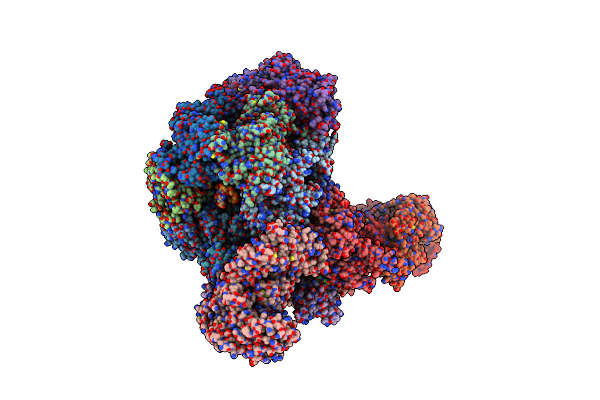 |
S. Cerevisiae Replisome-Scf(Dia2) Complex Bound To Double-Stranded Dna (Conformation Ii)
Organism: Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-11-10 Classification: REPLICATION Ligands: ANP, MG, ZN |
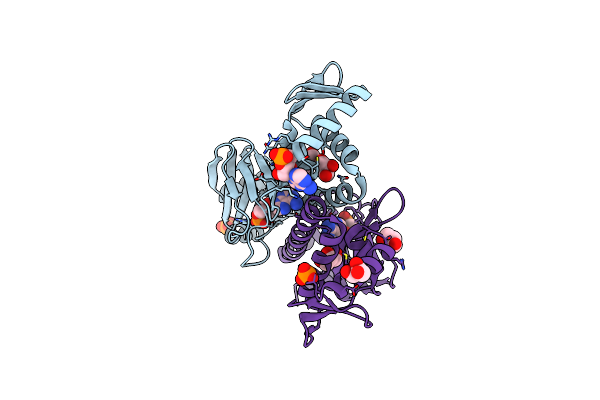 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.48 Å Release Date: 2013-10-30 Classification: TRANSCRIPTION Ligands: CMP, PO4, GOL |
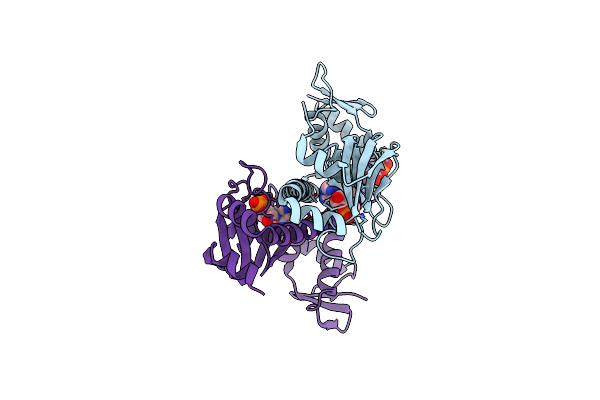 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-10-30 Classification: TRANSCRIPTION Ligands: CMP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2013-10-30 Classification: TRANSCRIPTION Ligands: CMP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2013-10-30 Classification: TRANSCRIPTION Ligands: CMP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2013-10-30 Classification: TRANSCRIPTION Ligands: CMP, GOL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-10-30 Classification: TRANSCRIPTION Ligands: CMP |
 |
Human Aldose Reductase In Complex With Nadp+ And The Inhibitor Lidorestat At 1.04 Angstrom
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.04 Å Release Date: 2006-03-14 Classification: OXIDOREDUCTASE Ligands: NDP, 3NA |

