Search Count: 31
 |
Crystal Structure Of A Ce15 From Fibrobacter Succinogenes Subsp. Succinogenes S85
Organism: Fibrobacter succinogenes subsp. succinogenes
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-08-28 Classification: HYDROLASE Ligands: ZN |
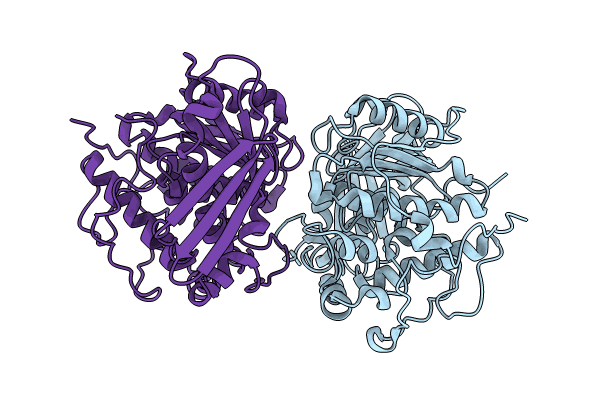 |
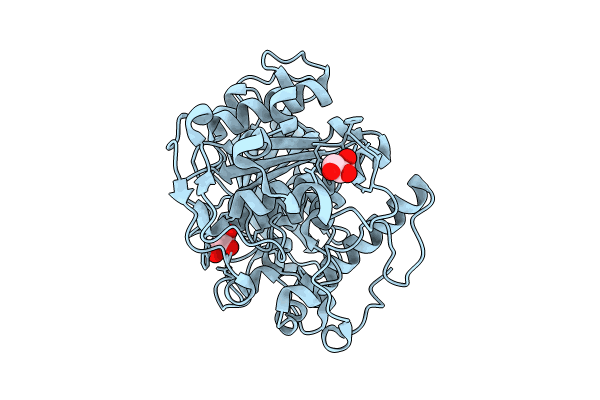
Crystal Structure Of A Ce15 Glucuronoyl Esterase From Piromyces Rhizinflatus
Organism: Piromyces rhizinflatus
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2024-08-28 Classification: HYDROLASE |
 |
Crystal Structure Of A Ce15 Glucuronoyl Esterase From Ruminococcus Flavefaciens
Organism: Ruminococcus flavefaciens 17
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2024-08-28 Classification: HYDROLASE Ligands: GOL |
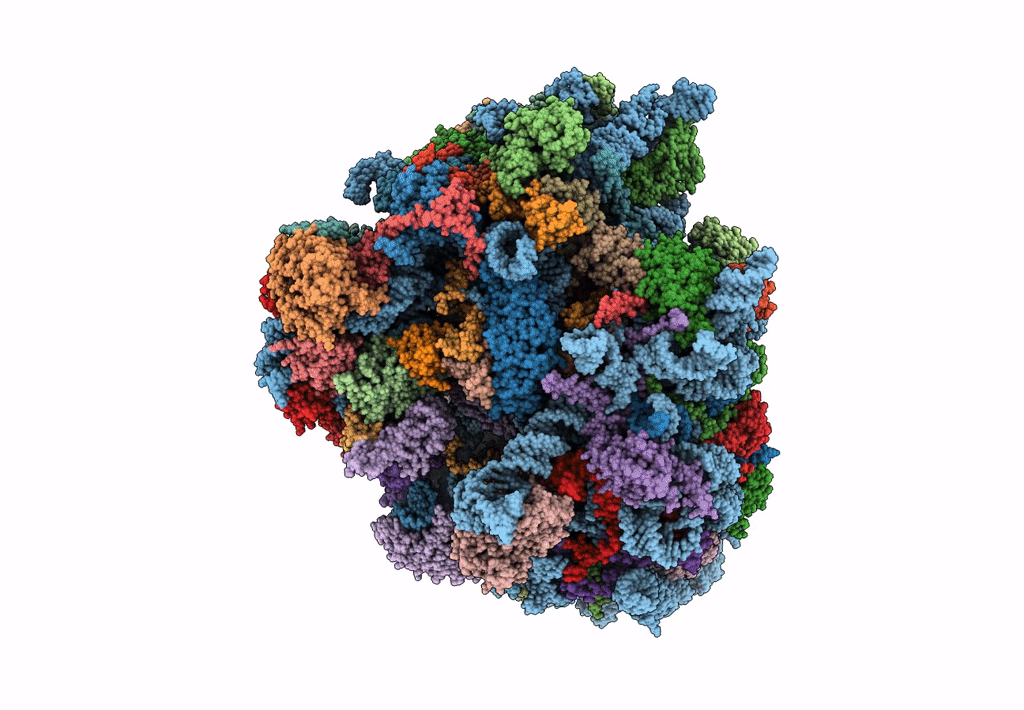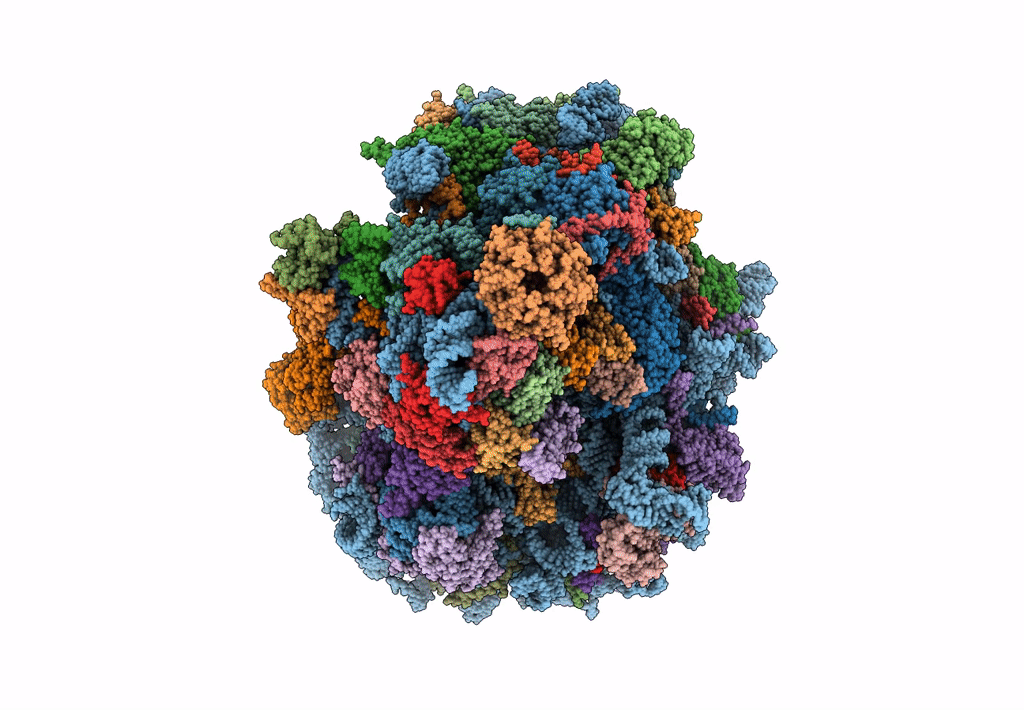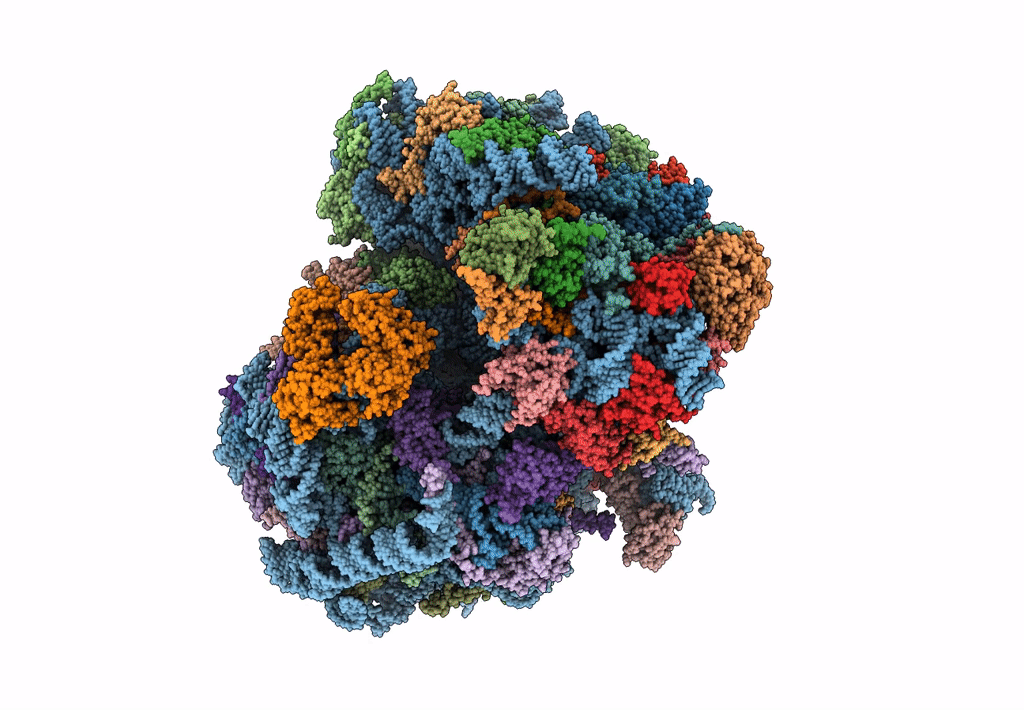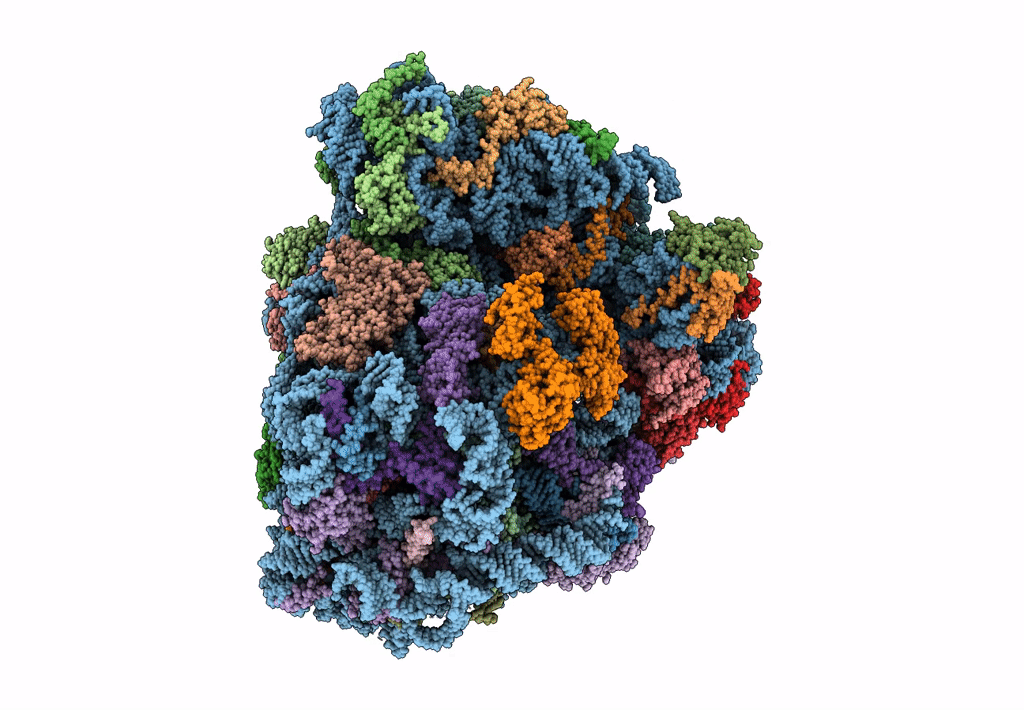 |
Cryo-Em Structure Of Stalled Rabbit 80S Ribosomes In Complex With Human Ccr4-Not And Cnot4
Organism: Homo sapiens, Oryctolagus cuniculus
Method: ELECTRON MICROSCOPY Release Date: 2023-09-06 Classification: RIBOSOME Ligands: MG, ZN |
 |
Structure Of Ampa Receptor Glua2 Complex With Auxiliary Subunits Tarp Gamma-5 And Cornichon-2 Bound To Competitive Antagonist Zk And Channel Blocker Spermidine (Closed State)
Organism: Rattus norvegicus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: ZK1, PCW, CLR, AJP, SPD |
 |
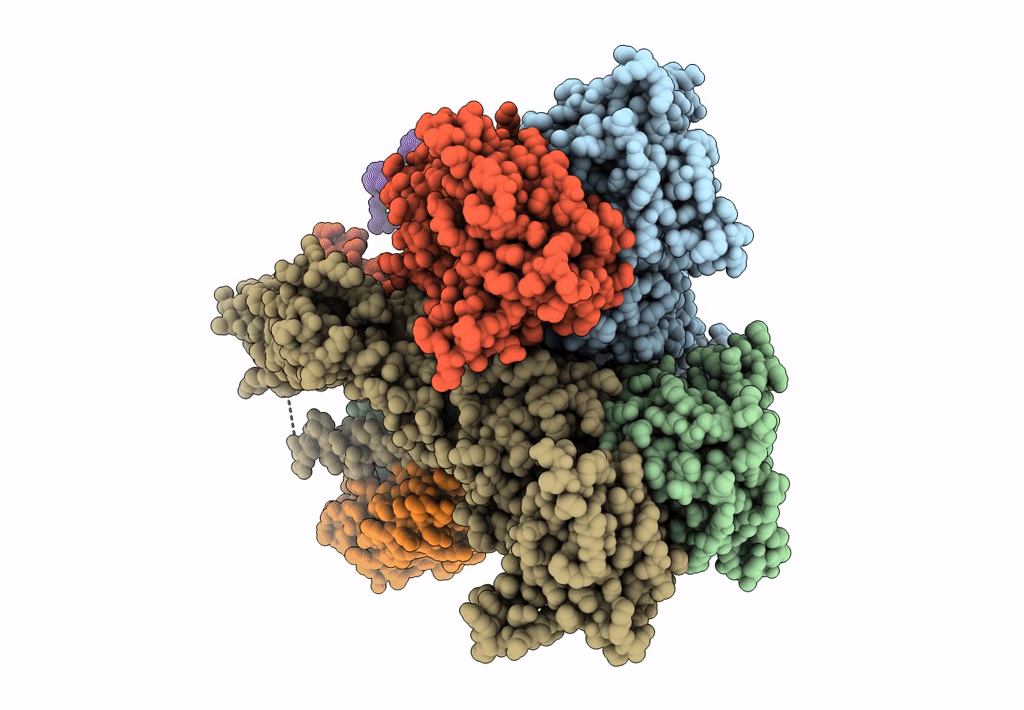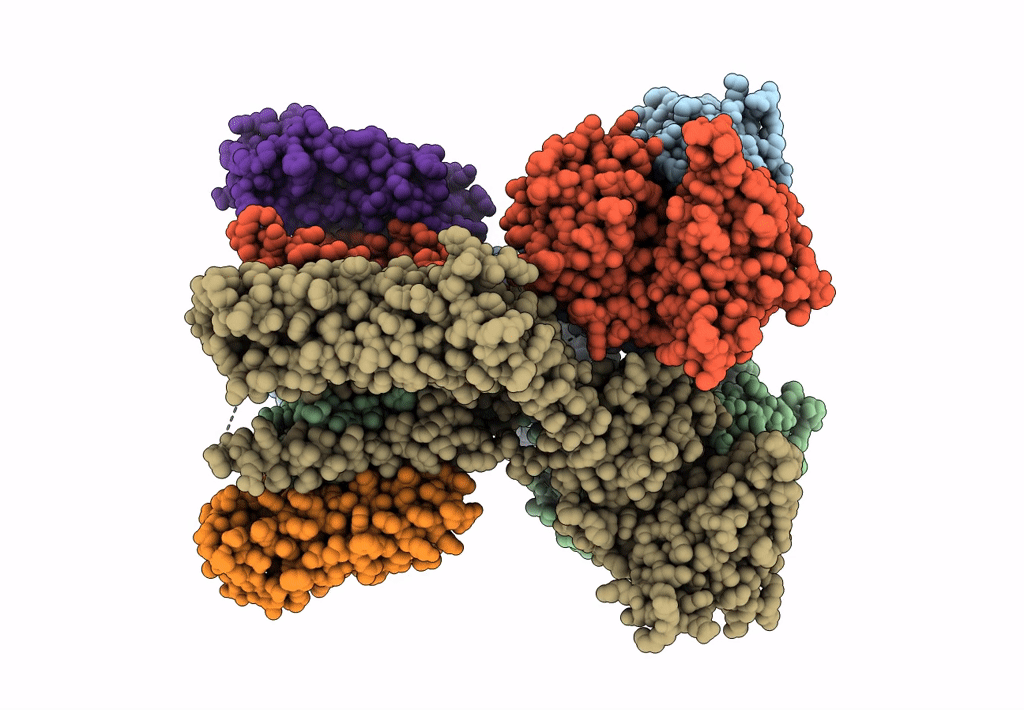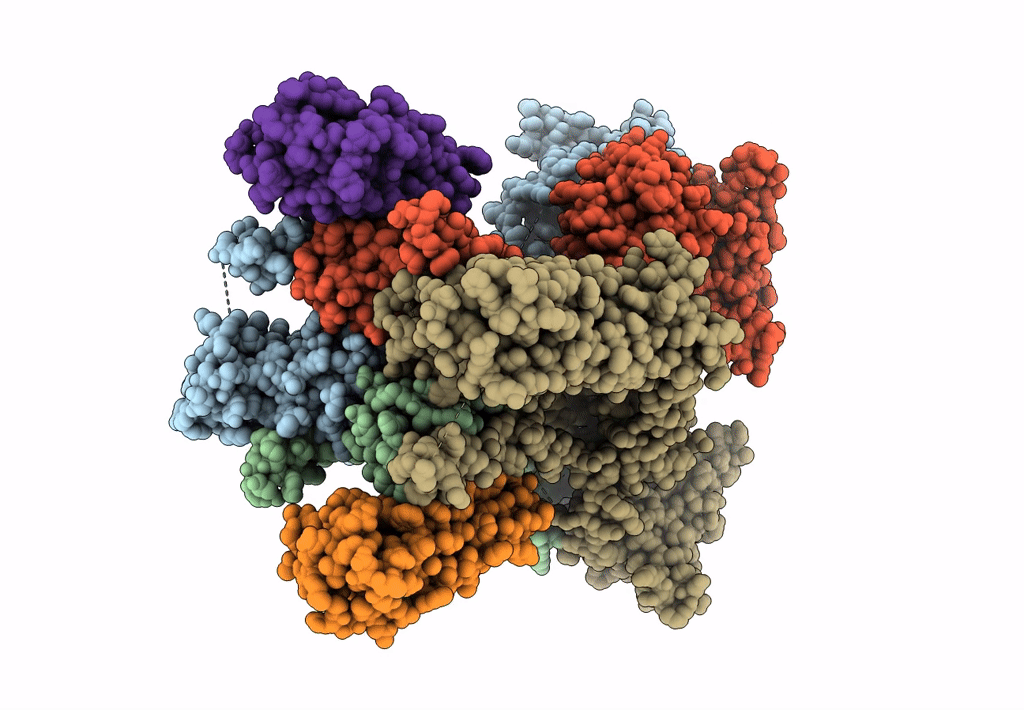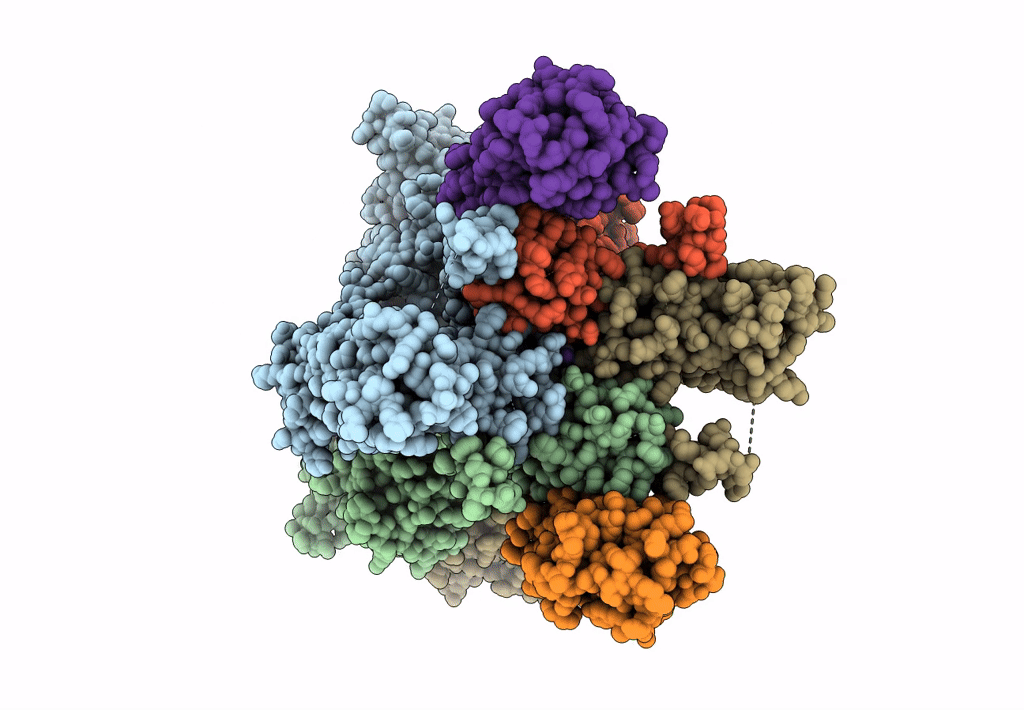
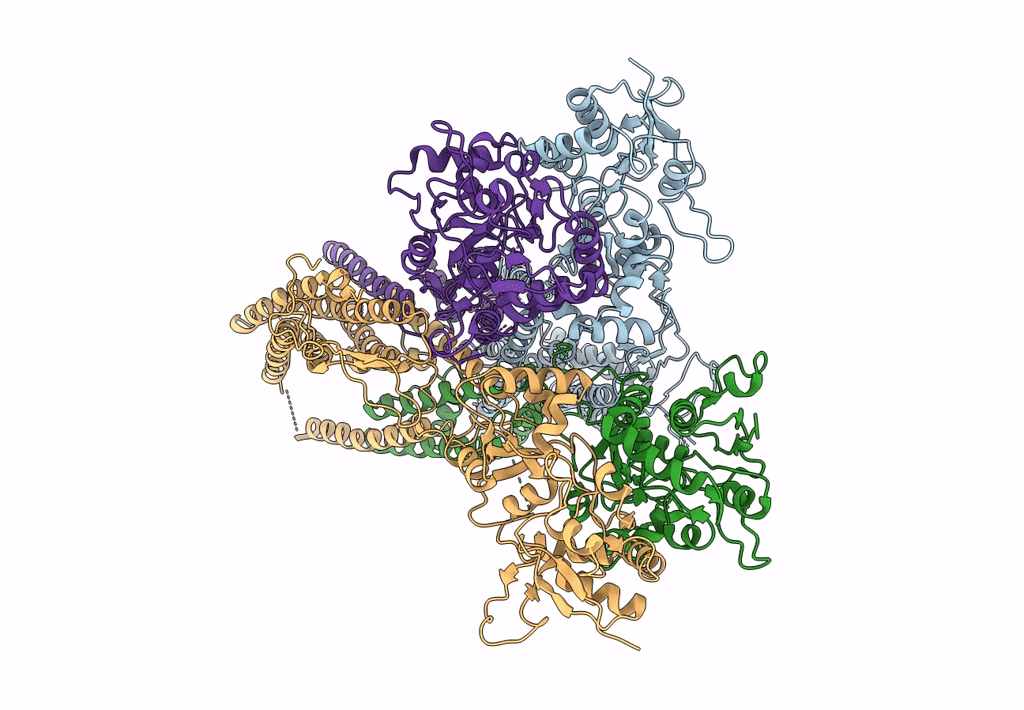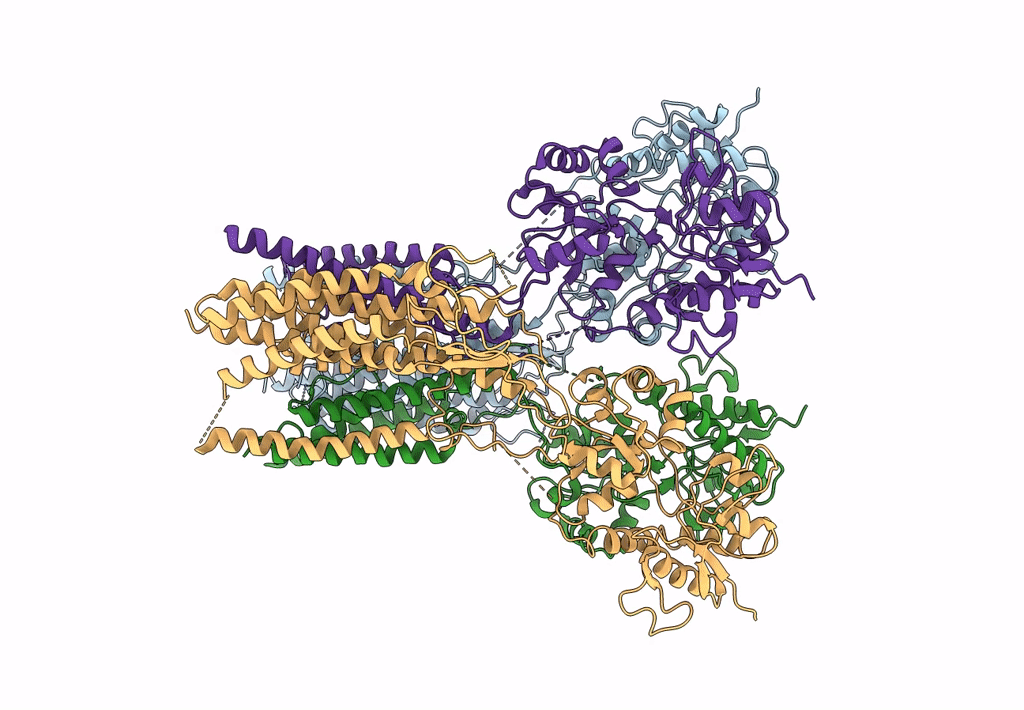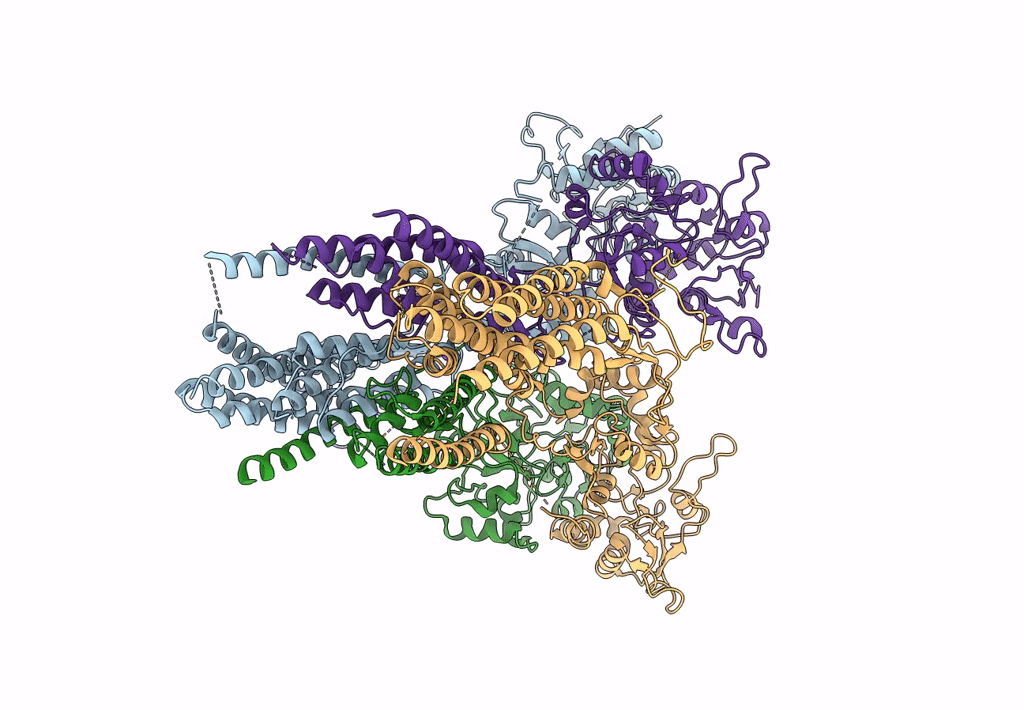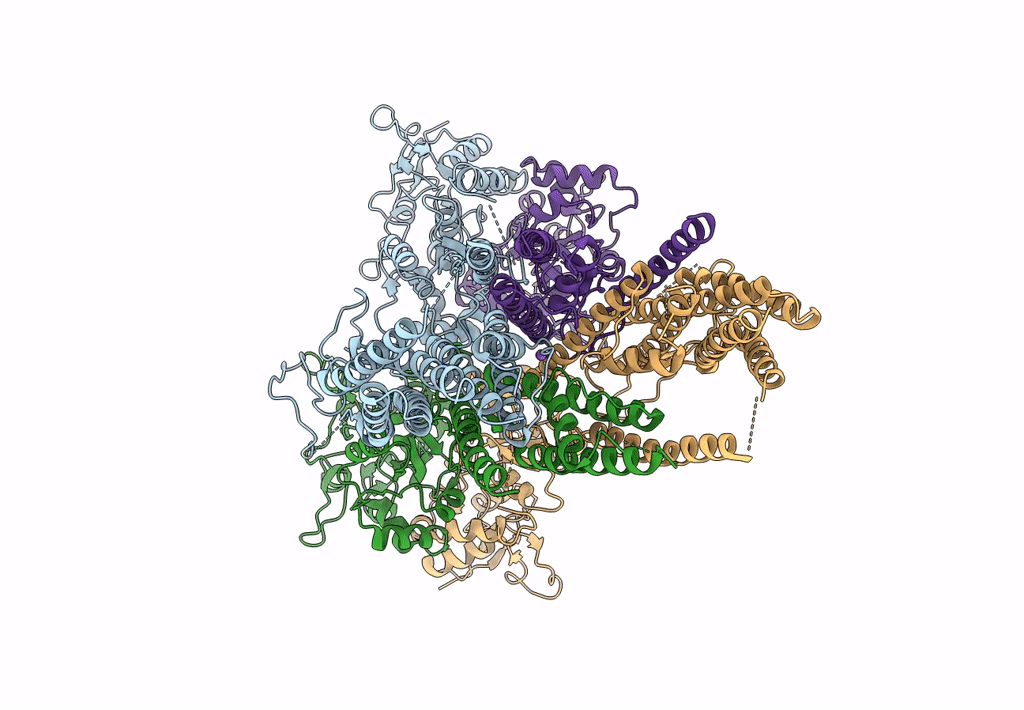
Structure Of Lbd-Tmd Of Ampa Receptor Glua2 In Complex With Auxiliary Subunits Tarp Gamma-5 And Cornichon-2 Bound To Competitive Antagonist Zk And Channel Blocker Spermidine (Closed State)
Organism: Rattus norvegicus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-09-06 Classification: SIGNALING PROTEIN Ligands: ZK1, PCW, CLR, AJP, SPD |
 |
Structure Of Lbd-Tmd Of Ampa Receptor Glua2 In Complex With Auxiliary Subunits Tarp Gamma-5 And Cornichon-2 (Apo State)
Organism: Rattus norvegicus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-09-06 Classification: SIGNALING PROTEIN Ligands: SPD |
 |
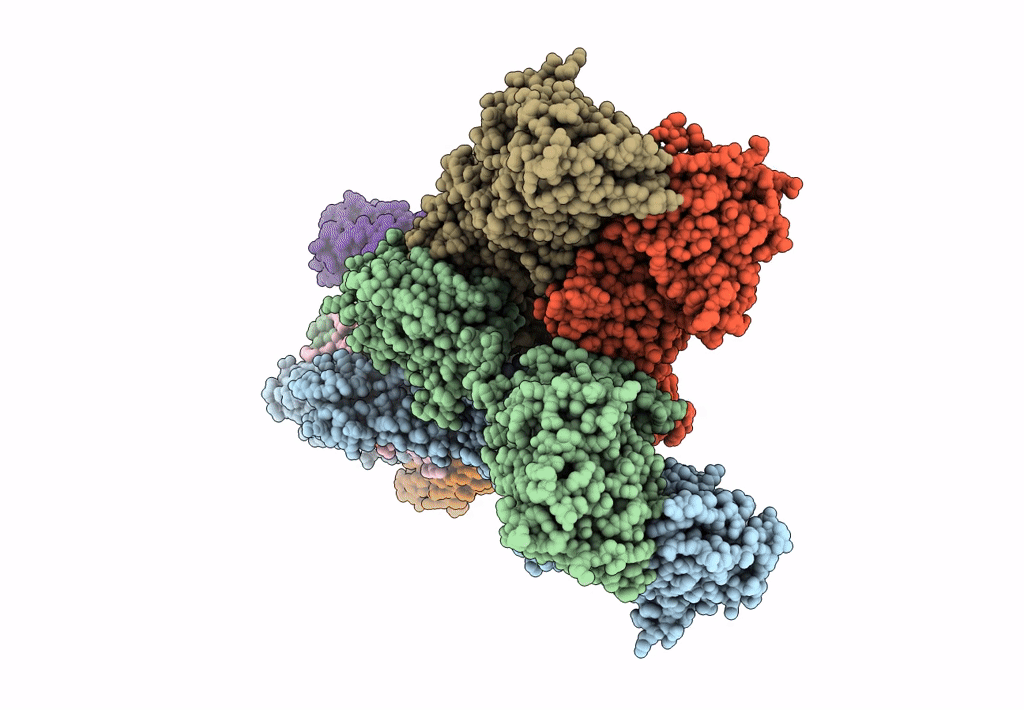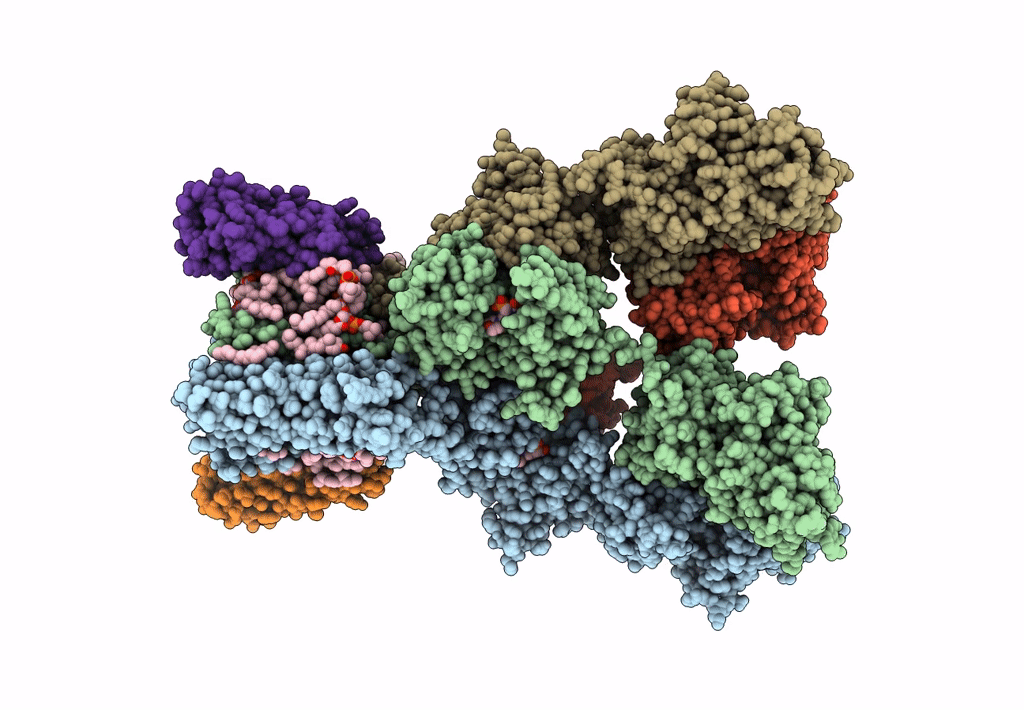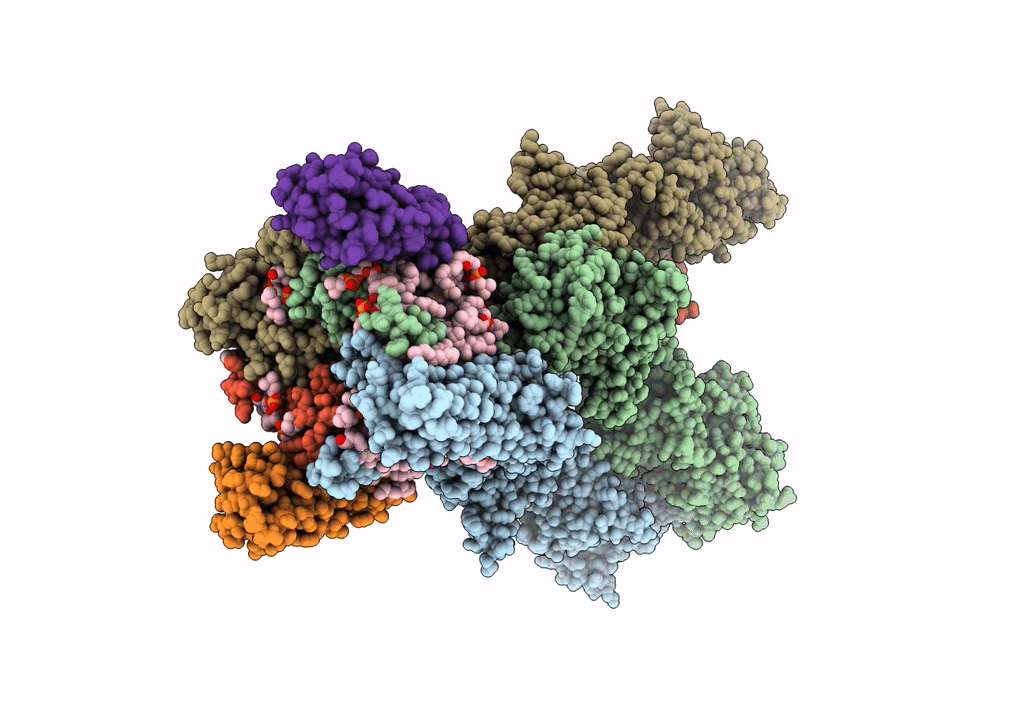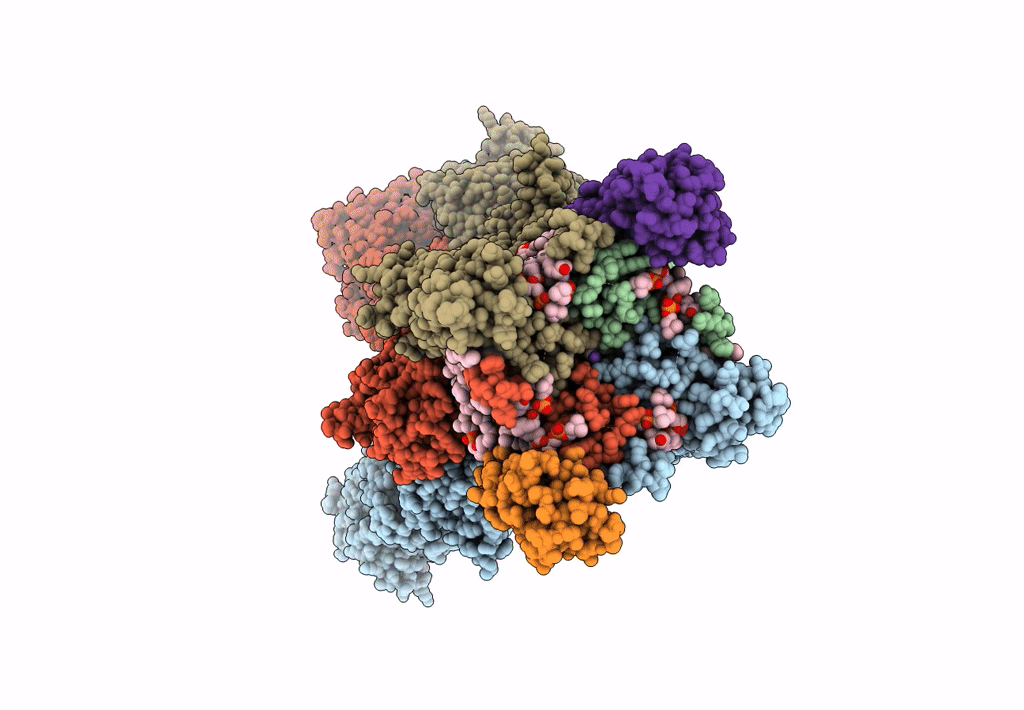
Structure Of Lbd-Tmd Of Ampa Receptor Glua2 In Complex With Auxiliary Subunit Tarp Gamma-5 (Apo State)
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2023-09-06 Classification: SIGNALING PROTEIN |
 |
Structure Of Ampa Receptor Glua2 Complex With Auxiliary Subunits Tarp Gamma-5 And Cornichon-2 Bound To Competitive Antagonist Zk, Channel Blocker Spermidine And Antiepileptic Drug Perampanel (Closed State)
Organism: Rattus norvegicus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-09-06 Classification: SIGNALING PROTEIN Ligands: 6ZP, ZK1, PCW, CLR, NA, SPD |
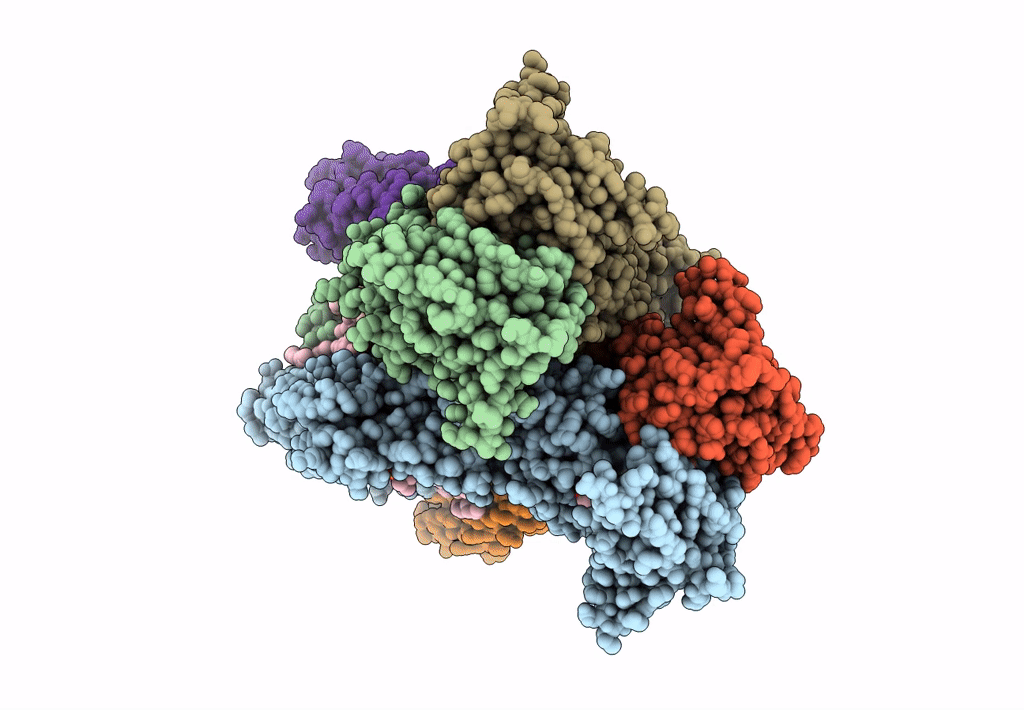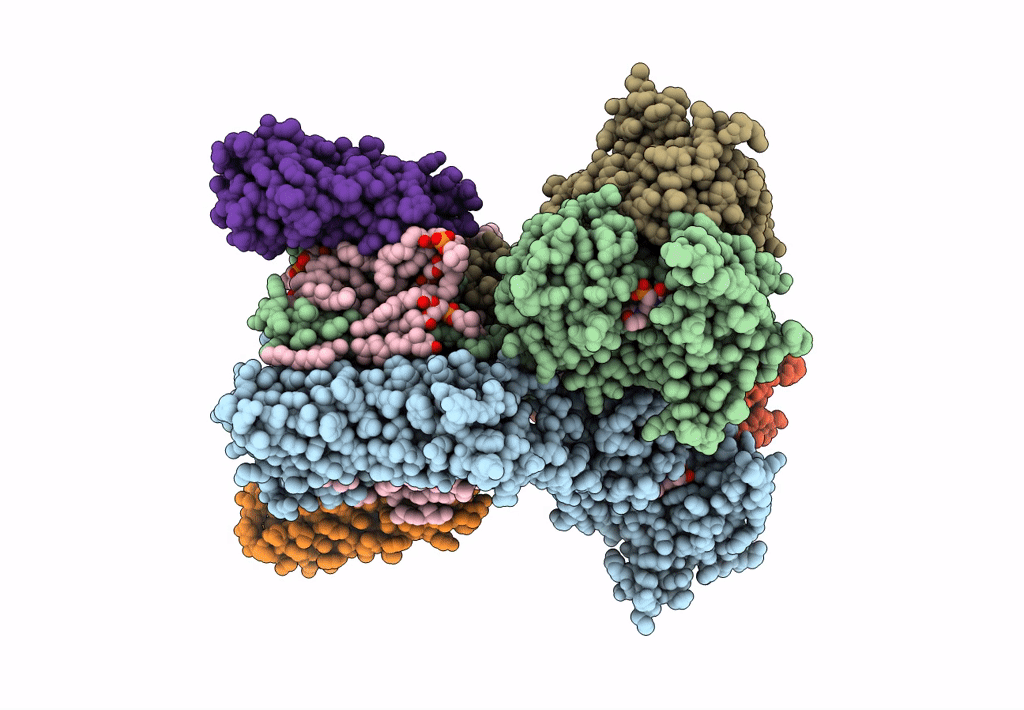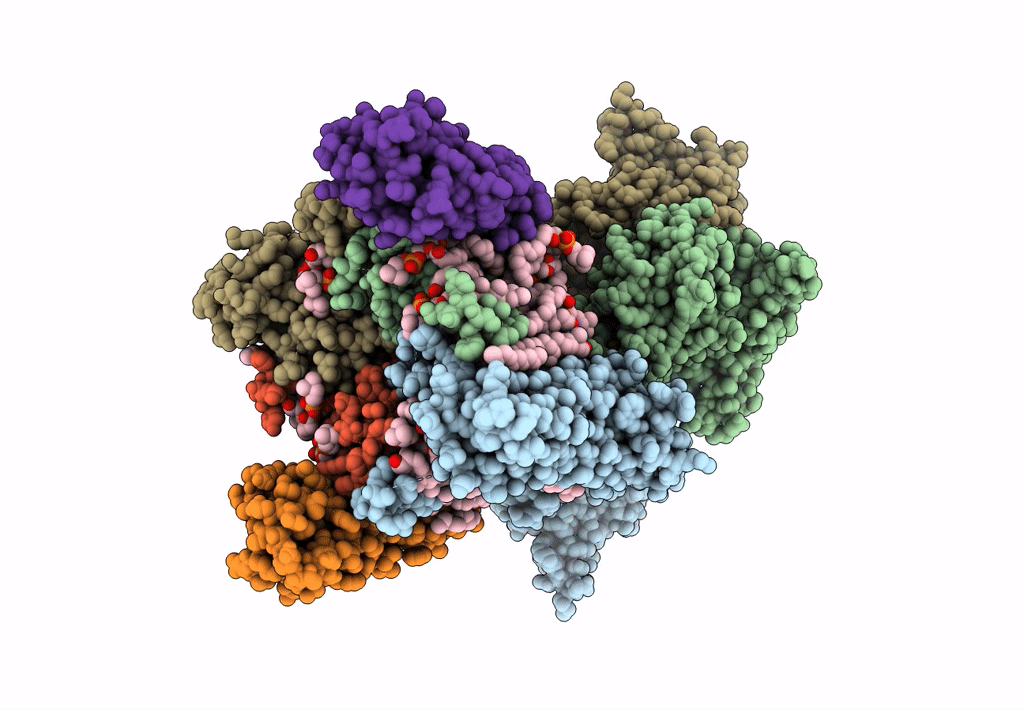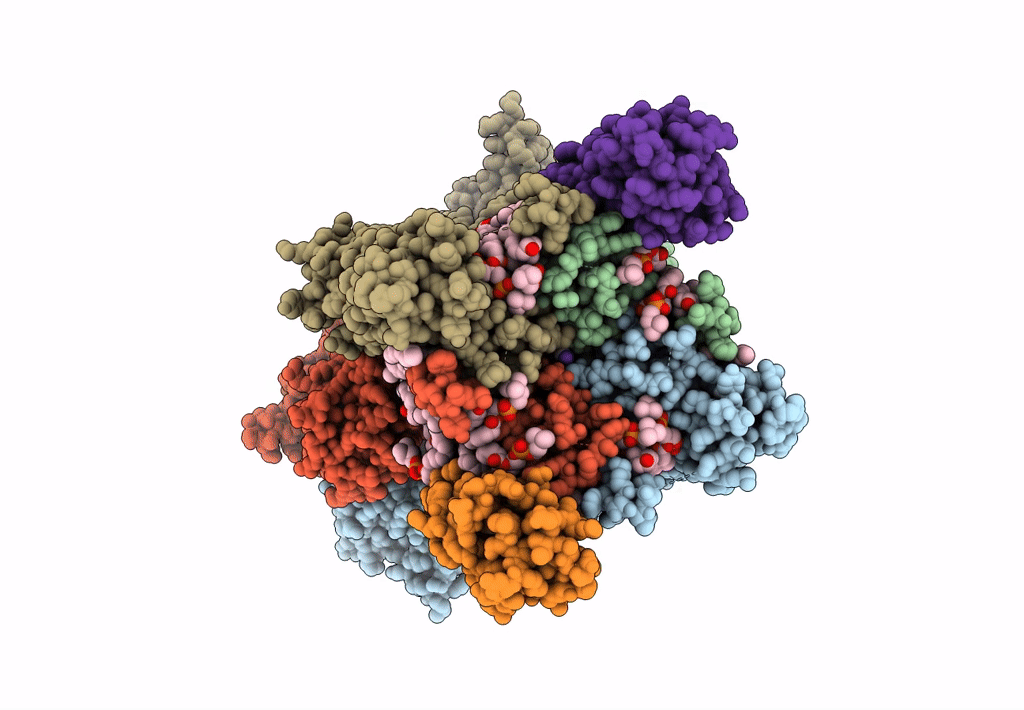 |
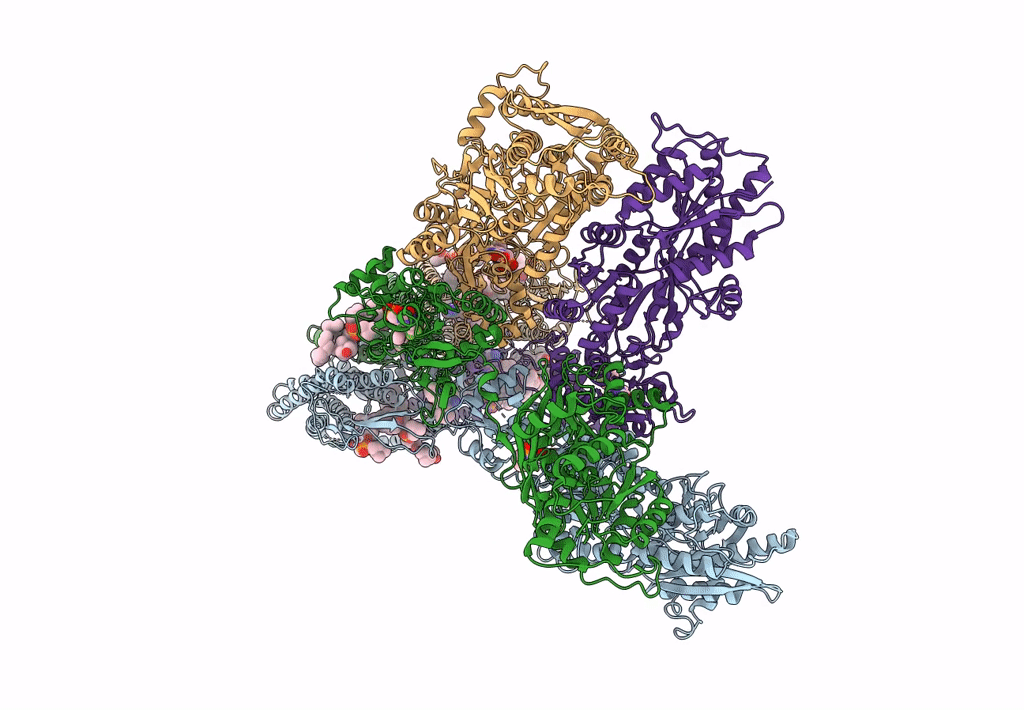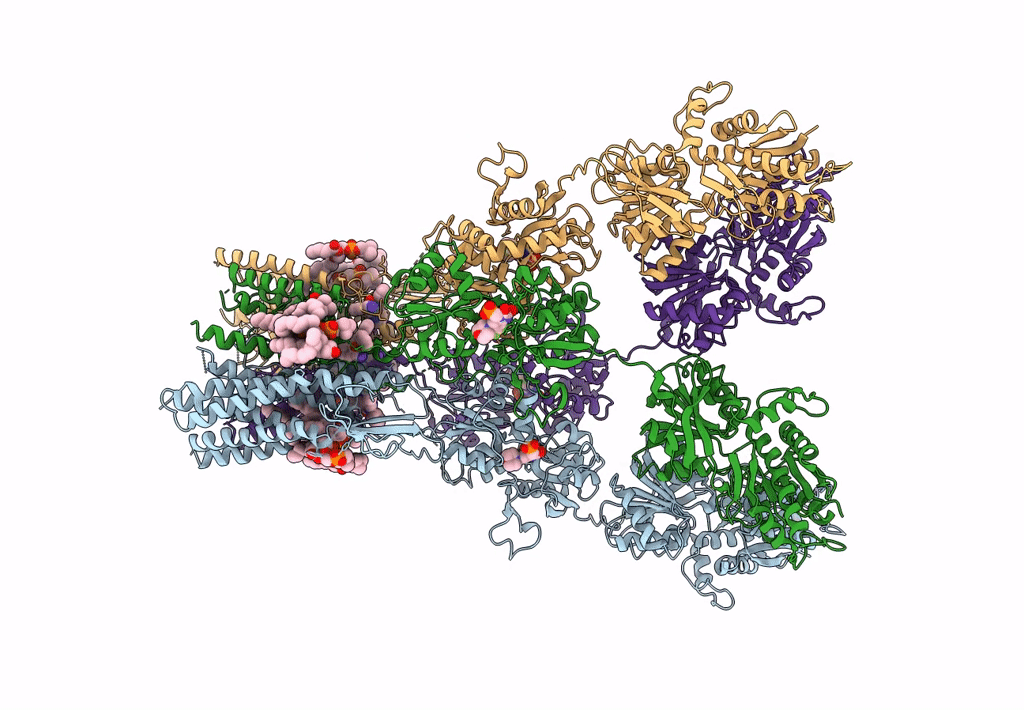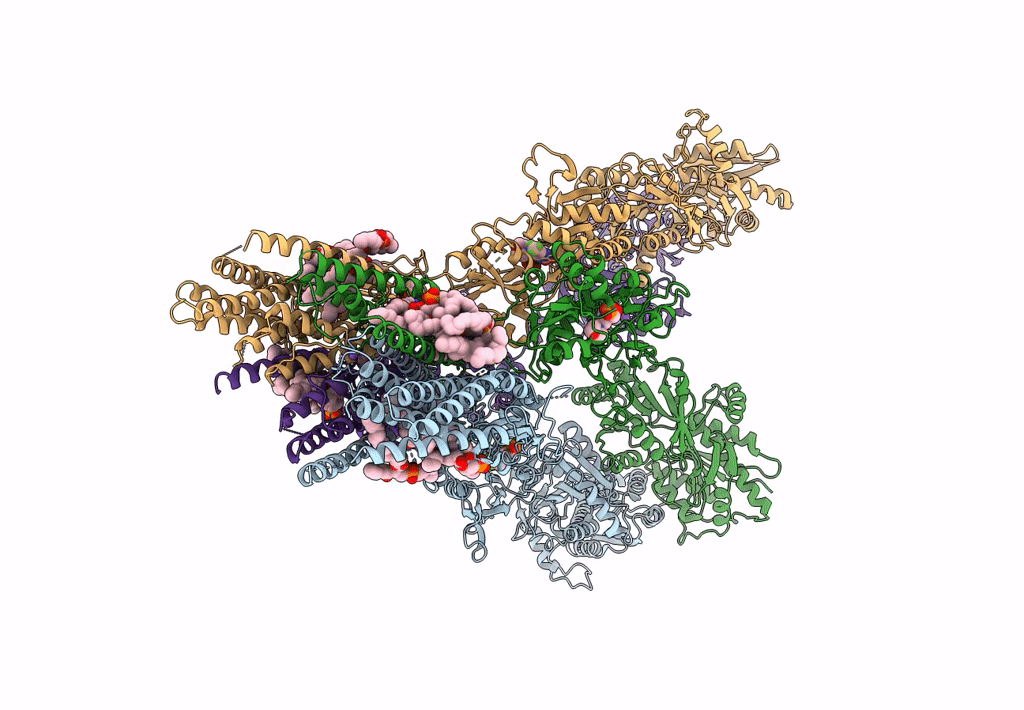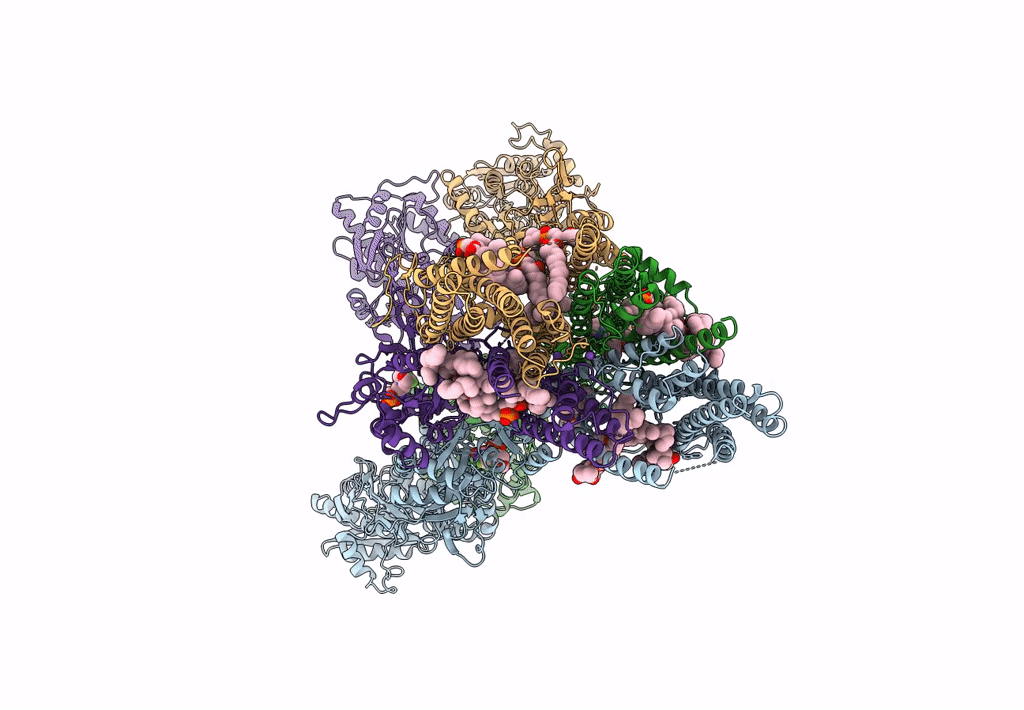
Structure Of Ampa Receptor Glua2 Complex With Auxiliary Subunits Tarp Gamma-5 And Cornichon-2 Bound To Competitive Antagonist Zk, Channel Blocker Spermidine And Antiepileptic Drug Perampanel (Closed State)
Organism: Rattus norvegicus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-09-06 Classification: SIGNALING PROTEIN Ligands: 6ZP, ZK1, PCW, CLR, NA, SPD |
 |
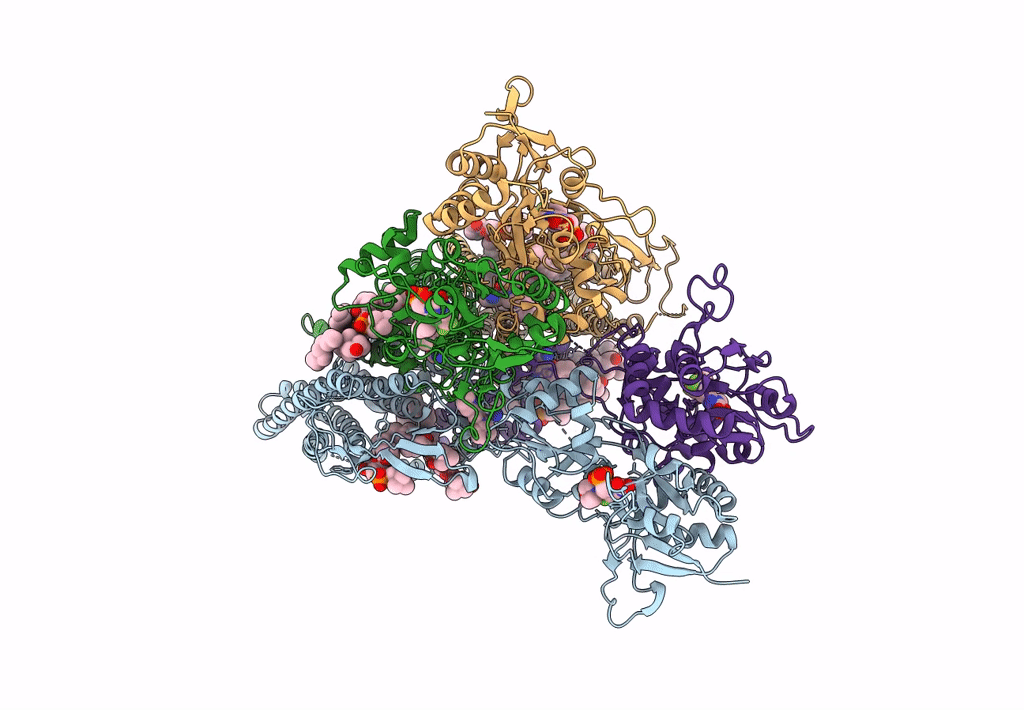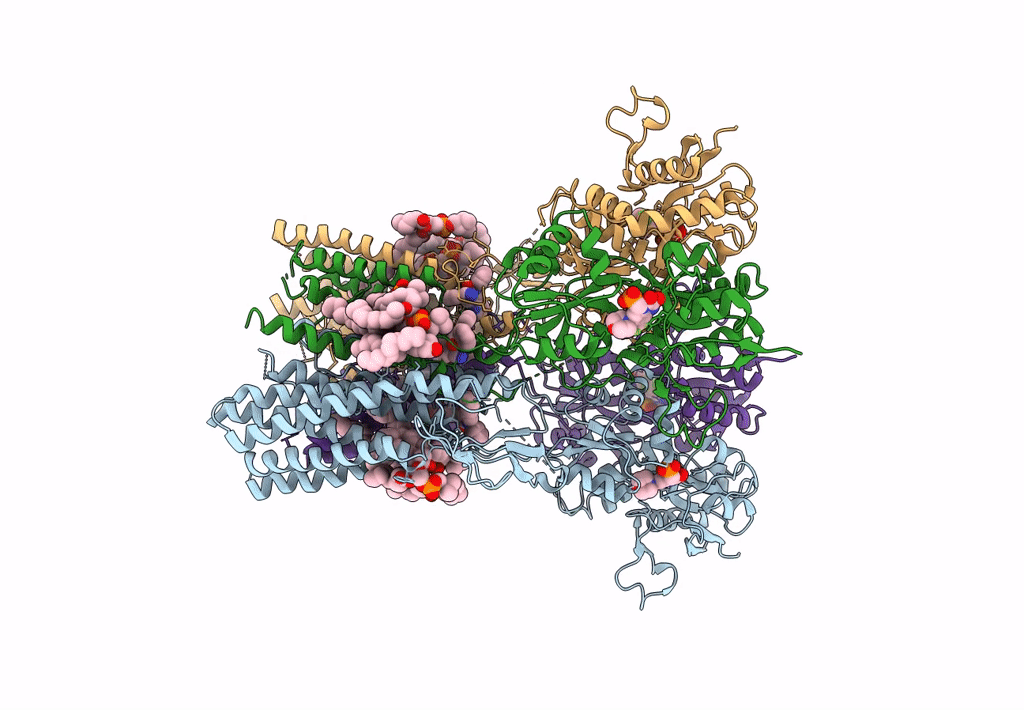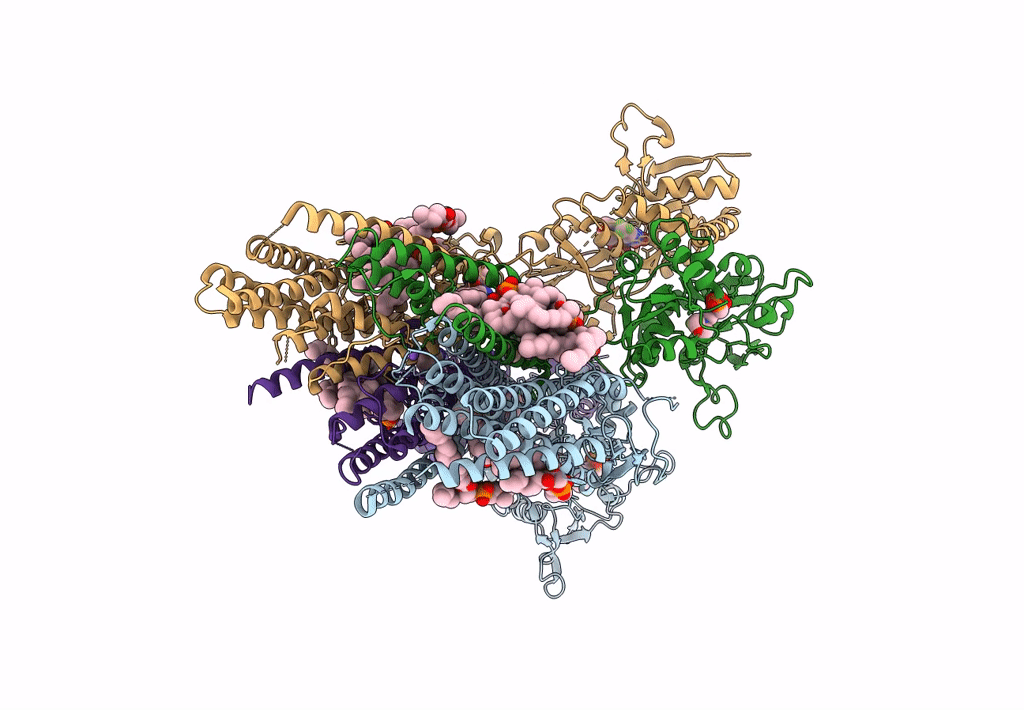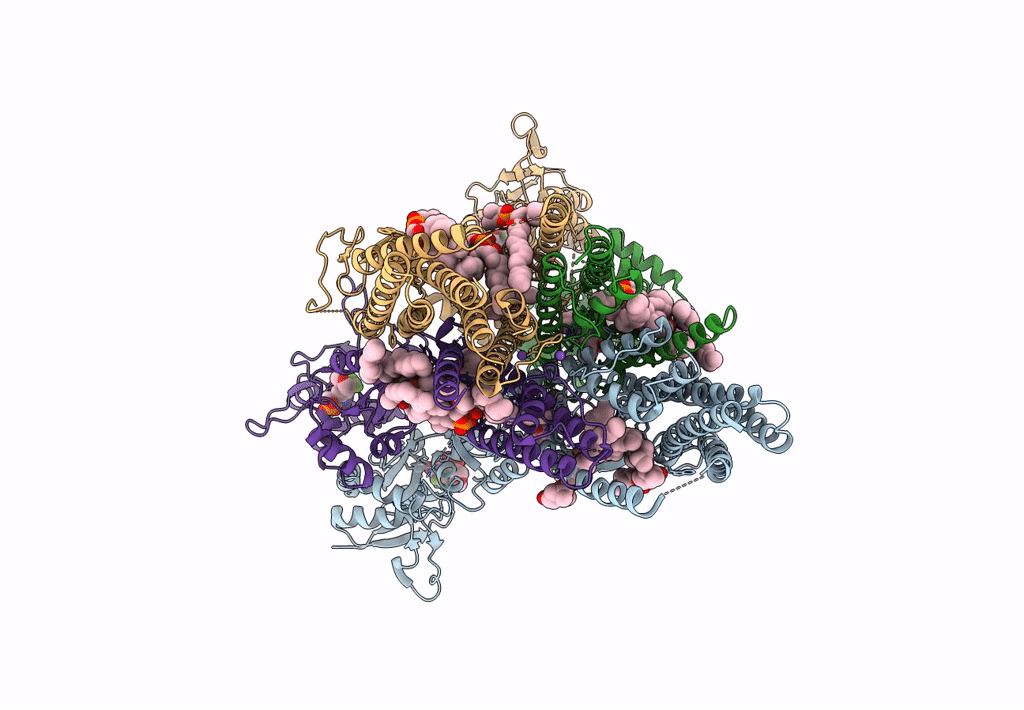
Structure Of Ampa Receptor Glua2 Complex With Auxiliary Subunit Tarp Gamma-5 Bound To Competitive Antagonist Zk And Antiepileptic Drug Perampanel (Closed State)
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2023-09-06 Classification: SIGNALING PROTEIN Ligands: 6ZP, ZK1, PCW, NA, CLR |
 |
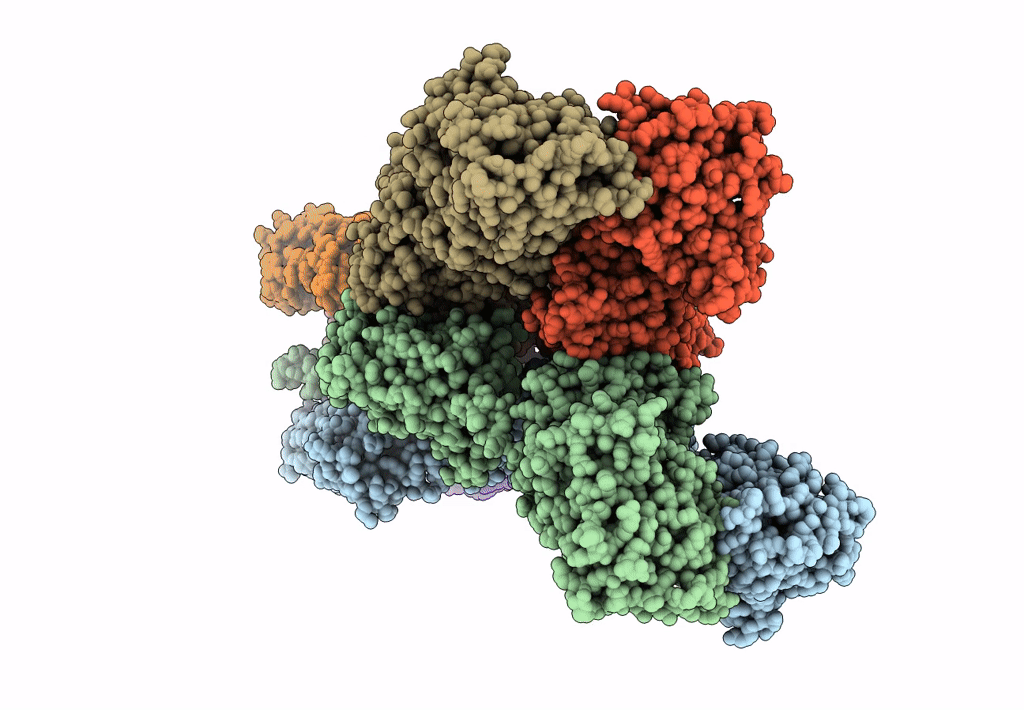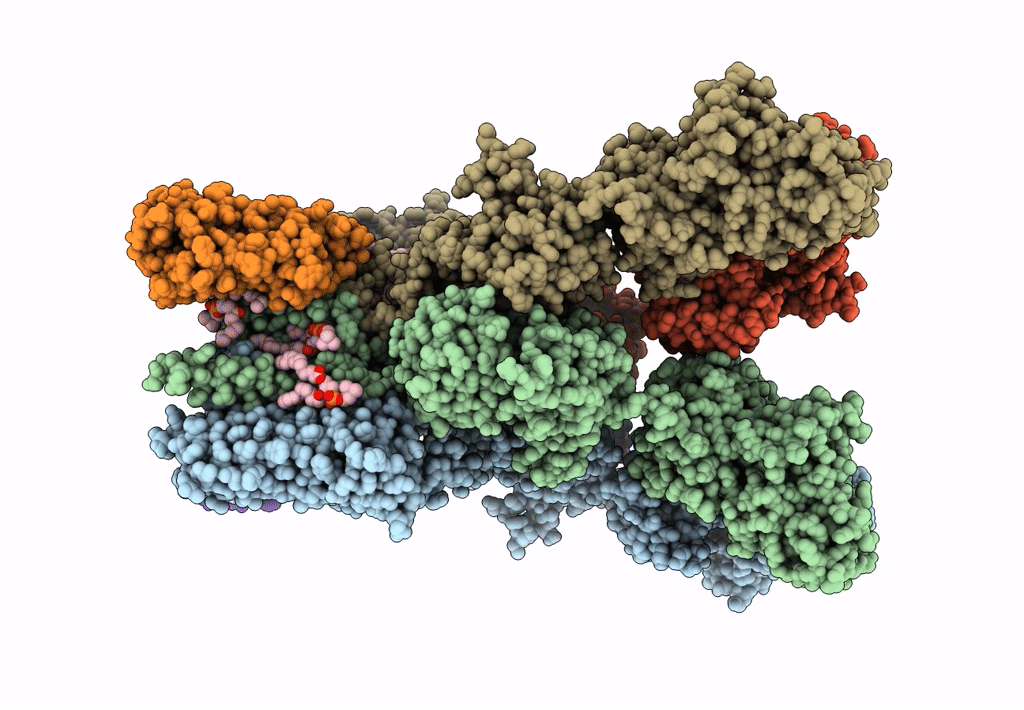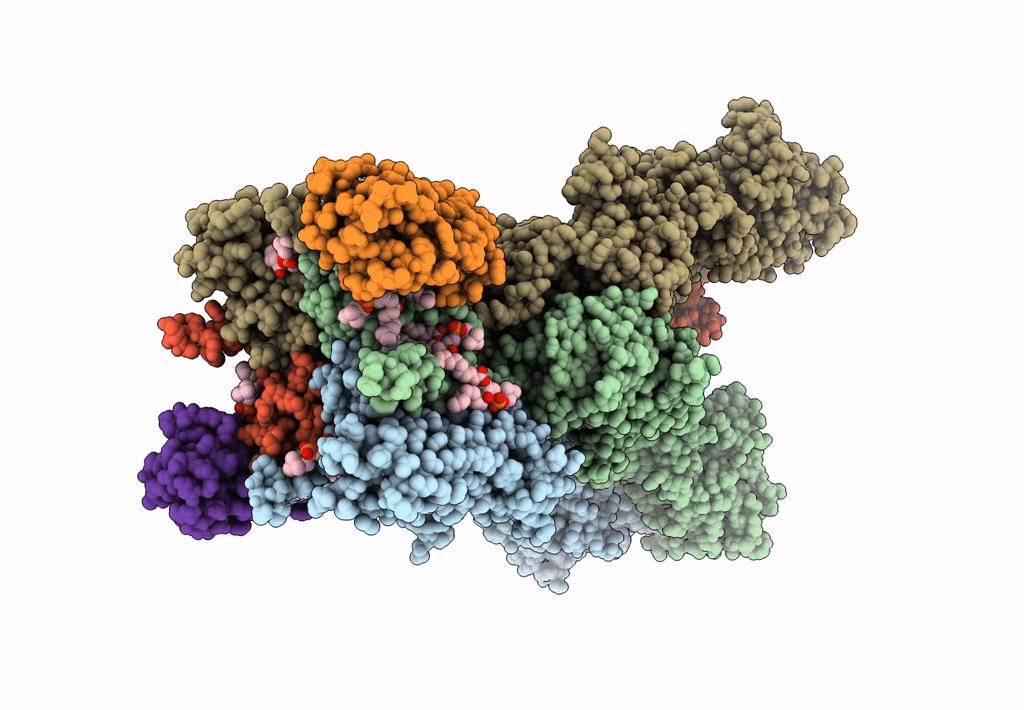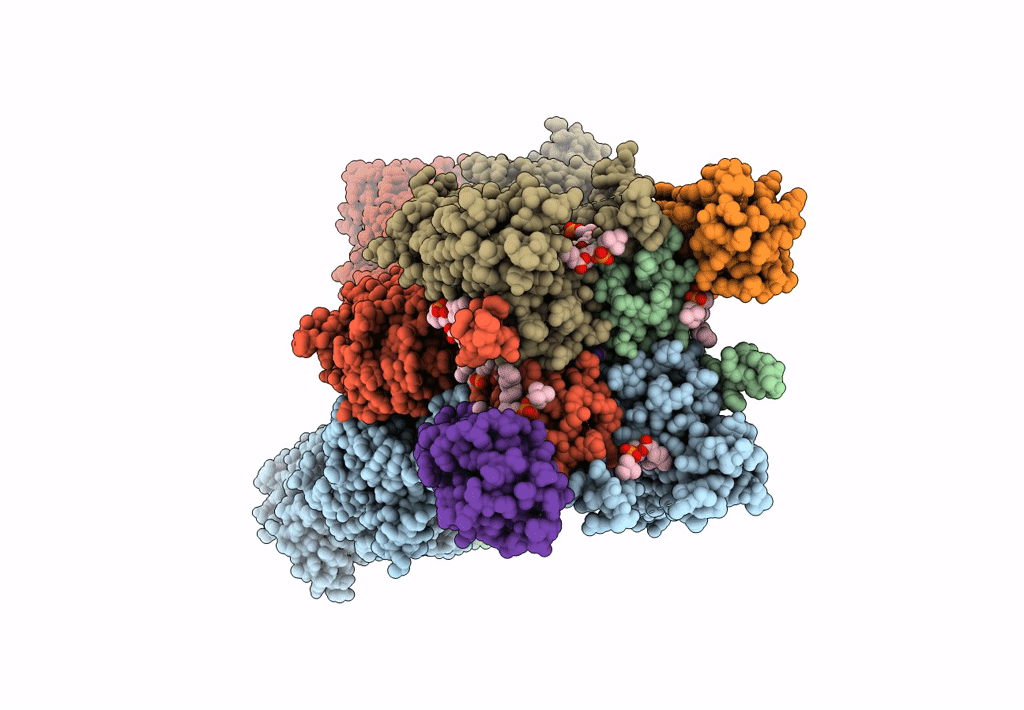
Structure Of Lbd-Tmd Of Ampa Receptor Glua2 In Complex With Auxiliary Subunit Tarp Gamma-5 Bound To Competitive Antagonist Zk And Antiepileptic Drug Perampanel (Closed State)
Organism: Rattus norvegicus
Method: ELECTRON MICROSCOPY Release Date: 2023-09-06 Classification: SIGNALING PROTEIN Ligands: 6ZP, ZK1, PCW, CLR, NA |
 |
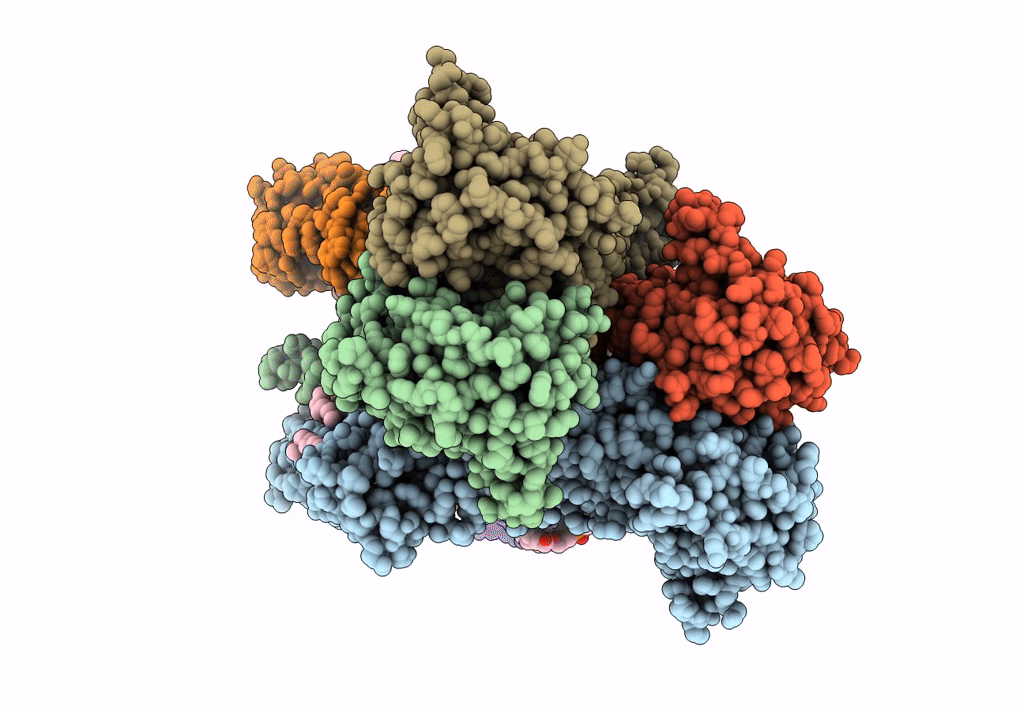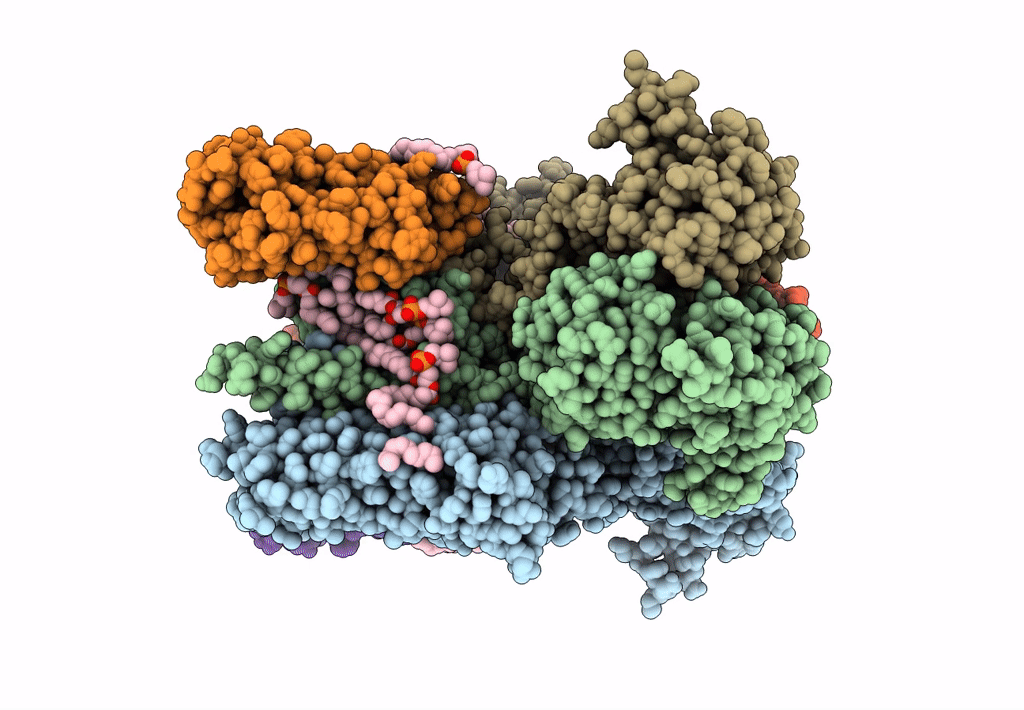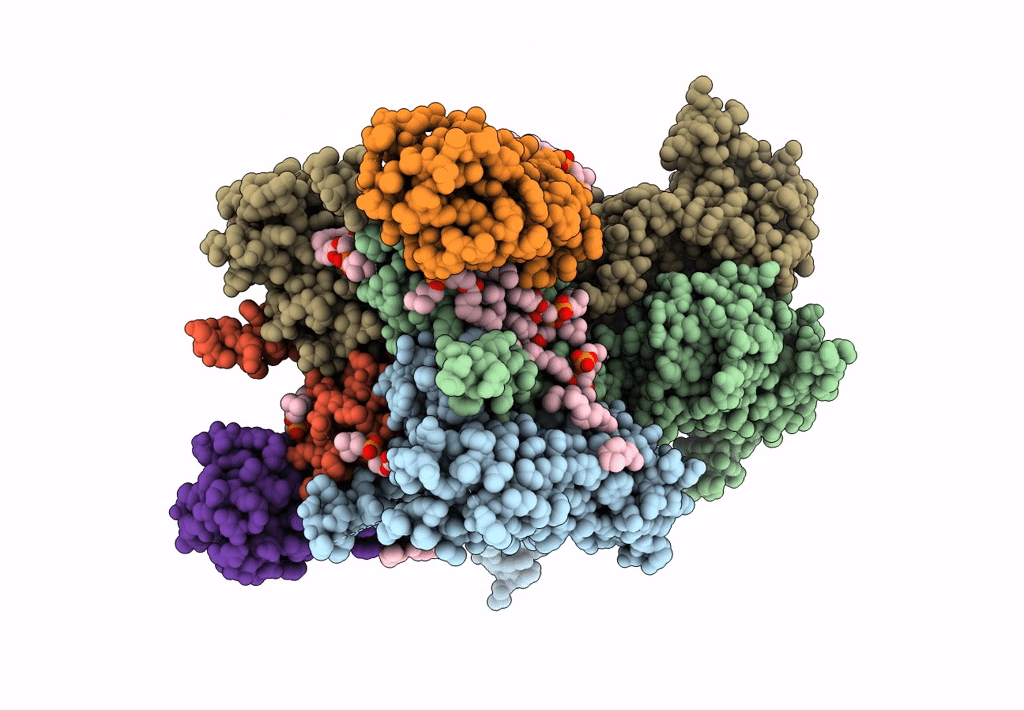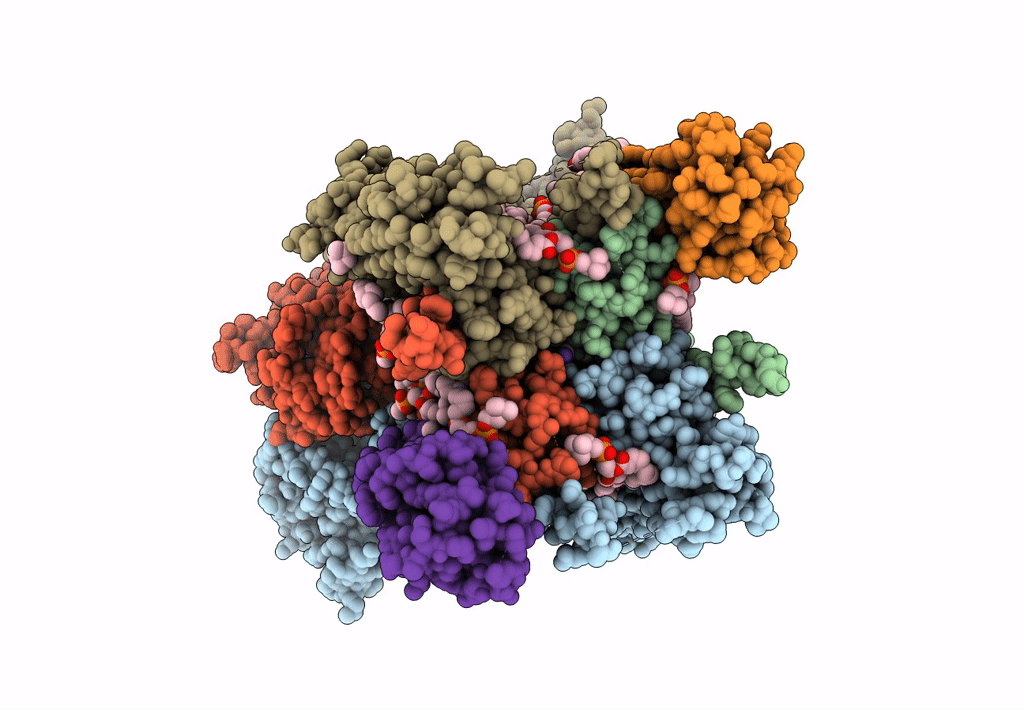
Structure Of Ampa Receptor Glua2 Complex With Auxiliary Subunits Tarp Gamma-5 And Cornichon-2 Bound To Glutamate And Channel Blocker Spermidine (Desensitized State)
Organism: Rattus norvegicus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-09-06 Classification: SIGNALING PROTEIN Ligands: PCW, GLU, SPD |
 |
Structure Of Lbd-Tmd Of Ampa Receptor Glua2 In Complex With Auxiliary Subunits Tarp Gamma-5 And Cornichon-2 Bound To Glutamate And Channel Blocker Spermidine (Desensitized State)
Organism: Rattus norvegicus, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-09-06 Classification: SIGNALING PROTEIN Ligands: PCW, GLU, SPD |
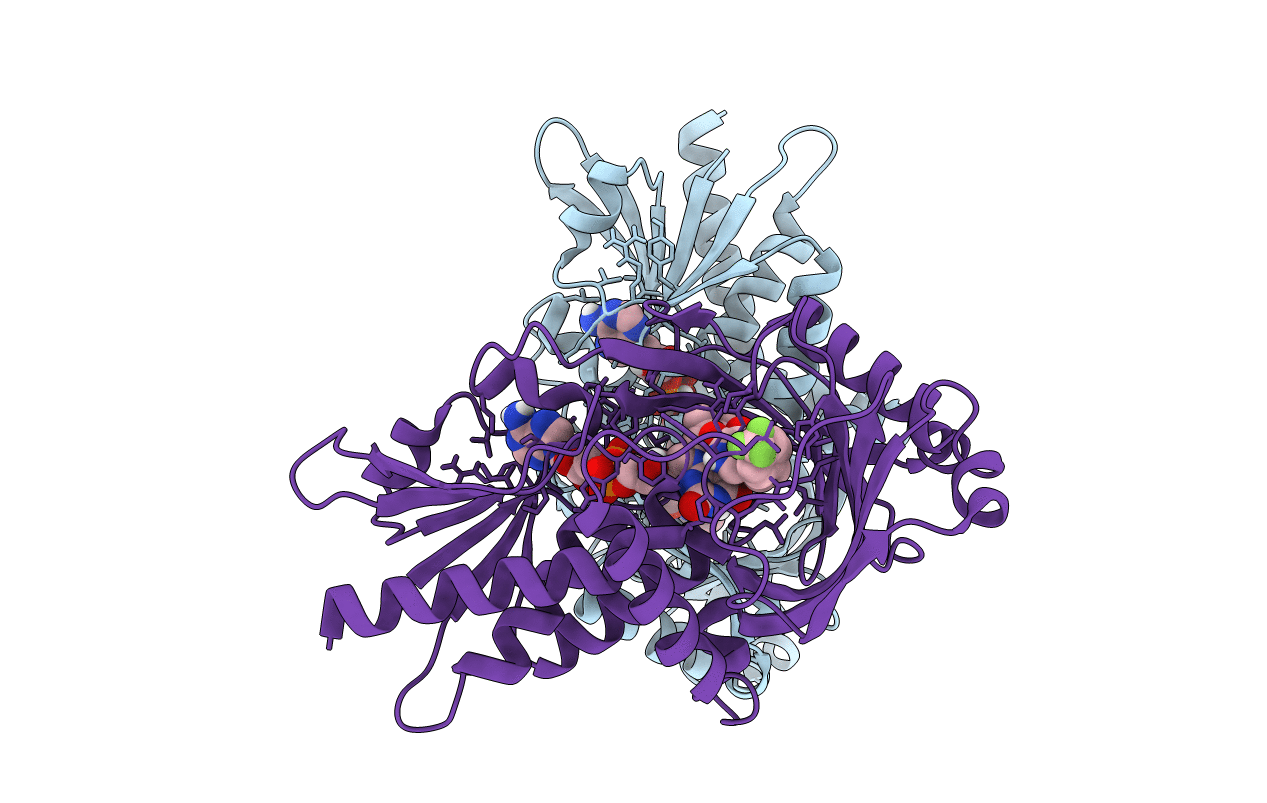 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-06-08 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: FAD, M7I |
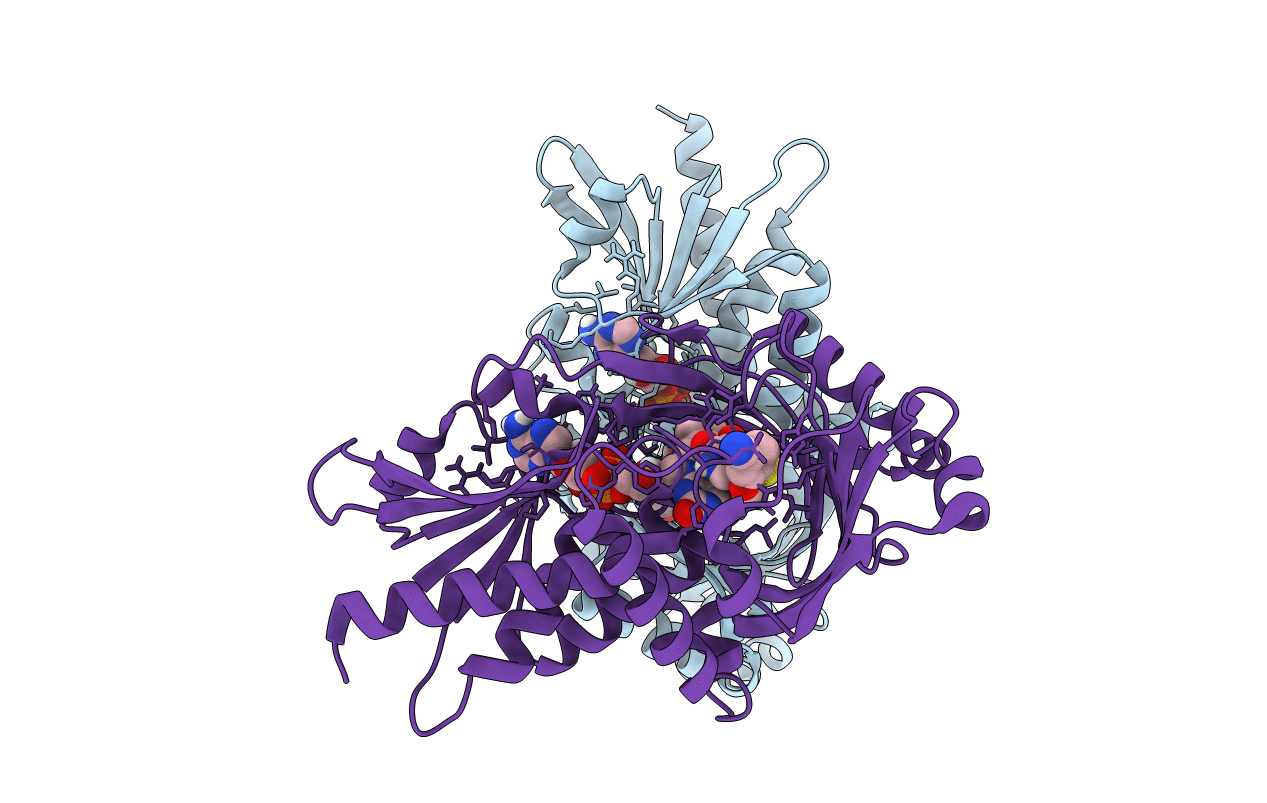 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2022-06-08 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: FAD, BEZ, M89 |
 |
Crystal Structure Of A Family 76 Glycoside Hydrolase From A Bovine Bacteroides Thetaiotaomicron Strain
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2020-04-29 Classification: HYDROLASE Ligands: P6G, EDO |
 |
Organism: Yersinia enterocolitica
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-11-20 Classification: LYASE Ligands: EDO |
 |
Organism: Yersinia enterocolitica
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-11-20 Classification: LYASE Ligands: EDO |
 |
Organism: Neocallimastix frontalis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2017-06-14 Classification: HYDROLASE Ligands: BTB, EDO, IOD |

