Search Count: 35
 |
Structure Of Human Serum Albumin In Complex With Aristolochic Acid Ii At 1.9 A Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-06-26 Classification: LIPID BINDING PROTEIN Ligands: MYR, EDO, GOR |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-06-26 Classification: LIPID BINDING PROTEIN Ligands: MYR, EDO |
 |
Structure Of Human Serum Albumin In Complex With Aristolochic Acid At 1.9 A Resolution
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-06-26 Classification: TRANSPORT PROTEIN Ligands: MYR, EDO, GOQ |
 |
Structure Of Human Serum Albumin In Complex With Aristolochic Acid I At 1.9 A Resolution - Optimized
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-06-26 Classification: TRANSPORT PROTEIN Ligands: MYR, EDO, GOQ |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2016-07-20 Classification: TRANSFERASE Ligands: ZN, CL, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2016-07-20 Classification: TRANSFERASE Ligands: ZN, CL |
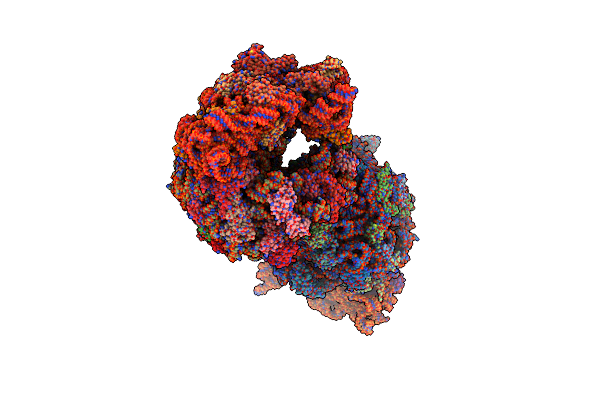 |
The Structure Of Thermorubin In Complex With The 70S Ribosome From Thermus Thermophilus.
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2014-07-09 Classification: RIBOSOME/antibiotic Ligands: T8B, MG |
 |
 |
 |
 |
Crystal Structure Of The Telomeric Saccharomyces Cerevisiae Cdc13 Ob2 Domain
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2012-11-28 Classification: CELL CYCLE |
 |
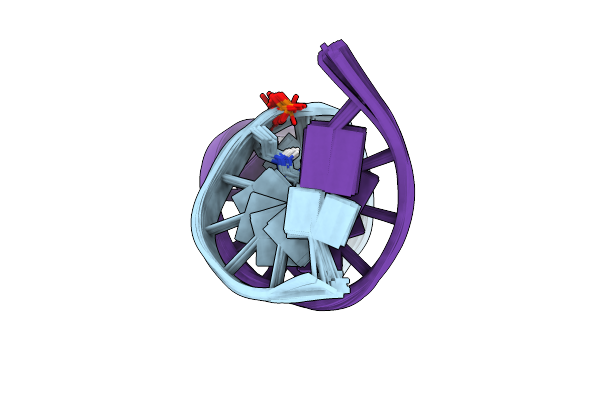
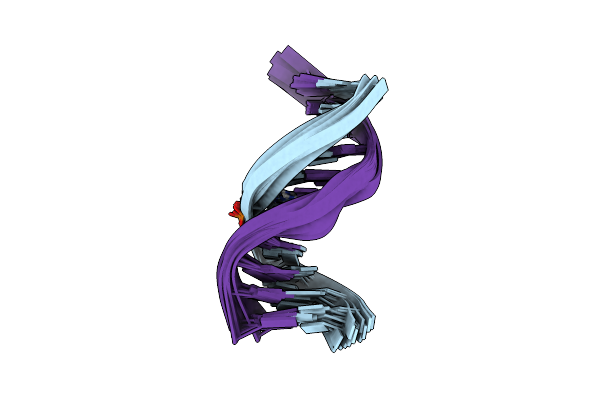
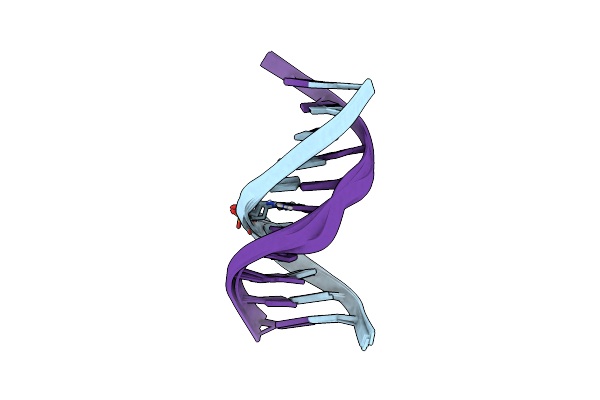
Nmr Structure Of Duplex Dna Containing The Alpha-Oh-Pdg Da Base Pair: A Mutagenic Intermediate Of Acrolein
|
 |
Nmr Structure Of Duplex Dna Containing The Beta-Oh-Pdg Da Base Pair: A Mutagenic Intermediate Of Acrolein
|
 |
 |
Crystal Structure Of Yeast Esa1 Hat Domain Complexed With H4K16Coa Bisubstrate Inhibitor
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-11-09 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: CMC |
 |
Crystal Structure Of Yeast Esa1 Hat Domain Bound To Coenzyme A With Active Site Lysine Acetylated
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-11-09 Classification: TRANSFERASE Ligands: COA, CAD, GOL, SO4 |
 |
Crystal Structure Of Yeast Esa1 E338Q Hat Domain Bound To Coenzyme A With Active Site Lysine Acetylated
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-11-09 Classification: TRANSFERASE Ligands: COA, EDO, SO4, CAD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2011-11-09 Classification: TRANSFERASE Ligands: ZN, CL, EDO |
 |
Human Mof E350Q Crystal Structure With Active Site Lysine Partially Acetylated
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2011-11-09 Classification: TRANSFERASE Ligands: ZN, CL |
 |
Crystal Structure Of The N-Terminal Domain Of The Yeast Telomere-Binding And Telomerase Regulatory Protein Cdc13
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-09-22 Classification: CELL CYCLE |

