Search Count: 45
 |
Organism: Homo sapiens, Plasmodium falciparum
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: IMMUNE SYSTEM/DE NOVO PROTEIN |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-04-09 Classification: LYASE Ligands: CL |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-04-09 Classification: LYASE |
 |
Crystal Structure Of Domain-Of-Unknown-Function Duf4867 From Bacillus Megaterium
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-02-26 Classification: ISOMERASE Ligands: FE, NA, CL |
 |
Crystal Structure Of Domain-Of-Unknown-Function Duf4867 From Bacillus Megaterium (Unmodelled Additional Ligand Density At Active Site)
Organism: Priestia megaterium
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-02-26 Classification: ISOMERASE Ligands: FE |
 |
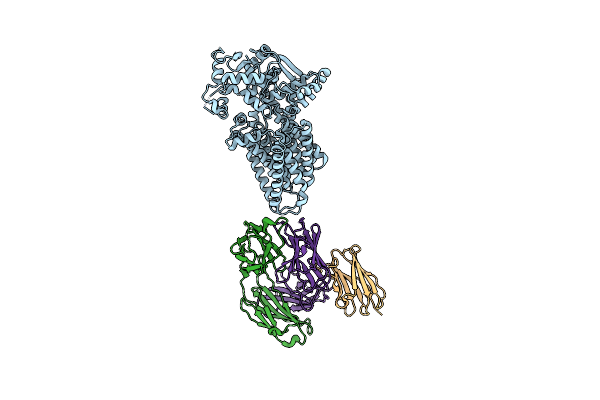
Cryo-Em Structure Of C-Mannosyltransferase Cedpy19, In Complex With Acceptor Peptide And Bound To Cmt2-Fab And Anti-Fab Nanobody
Organism: Synthetic construct, Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Structure Of C-Mannosyltransferase Cedpy19, In Apo State, Bound To Cmt2-Fab And Anti-Fab Nanobody
Organism: Synthetic construct, Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: MEMBRANE PROTEIN |
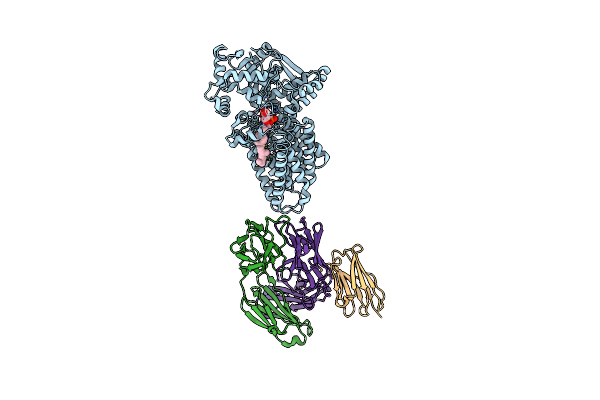 |
Cryo-Em Structure Of C-Mannosyltransferase Cedpy19, In Complex With Dol25-P-Man And Bound To Cmt2-Fab And Anti-Fab Nanobody
Organism: Synthetic construct, Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: MEMBRANE PROTEIN Ligands: IZY |
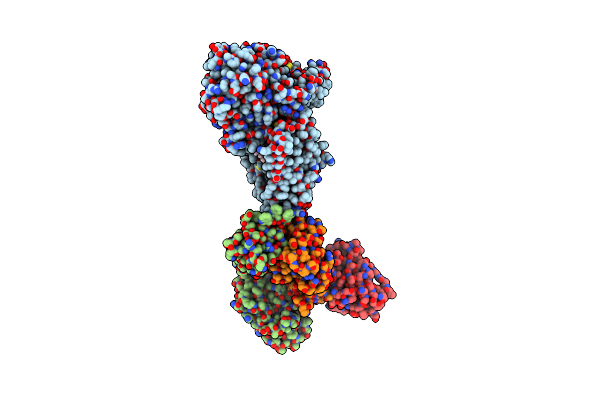 |
Cryo-Em Structure Of C-Mannosyltransferase Cedpy19, In Ternary Complex With Dol25-P-C-Man And Acceptor Peptide, Bound To Cmt2-Fab And Anti-Fab Nanobody
Organism: Synthetic construct, Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: MEMBRANE PROTEIN Ligands: IZU |
 |
Organism: Plasmodium falciparum (isolate 3d7)
Method: X-RAY DIFFRACTION Resolution:3.51 Å Release Date: 2022-03-30 Classification: HYDROLASE Ligands: MG |
 |
Streptococcal High Identity Repeats In Tandem (Shirt) Domains 3-4 From Cell Surface Protein Sgo_0707
Organism: Streptococcus gordonii (strain challis / atcc 35105 / bcrc 15272 / ch1 / dl1 / v288)
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2021-06-09 Classification: CELL ADHESION Ligands: CL, TRS |
 |
Streptococcal High Identity Repeats In Tandem (Shirt) Domain 2 From Cell Surface Protein Sgo_0707
Organism: Streptococcus gordonii (strain challis / atcc 35105 / bcrc 15272 / ch1 / dl1 / v288)
Method: X-RAY DIFFRACTION Resolution:0.95 Å Release Date: 2021-06-09 Classification: CELL ADHESION |
 |
Streptococcal High Identity Repeats In Tandem (Shirt) Domain 10 From Cell Surface Protein Sgo_0707
Organism: Streptococcus gordonii (strain challis / atcc 35105 / bcrc 15272 / ch1 / dl1 / v288)
Method: X-RAY DIFFRACTION Resolution:0.82 Å Release Date: 2021-06-09 Classification: CELL ADHESION Ligands: IS8 |
 |
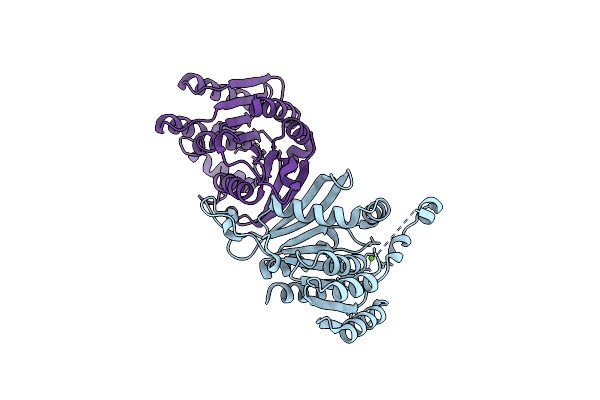
Crystal Structure Of Hydroxyacyl Glutathione Hydrolase (Glob) From Staphylococcus Aureus, Apoenzyme
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-12-30 Classification: HYDROLASE Ligands: ZN, SO4 |
 |
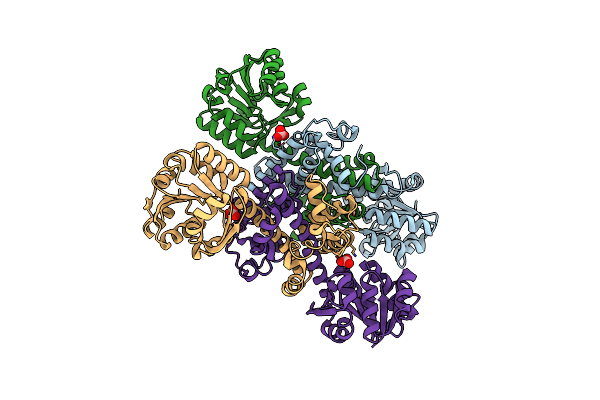
Crystal Structure Of S-Formylglutathione Hydrolase (Frmb) From Staphylococcus Aureus, Apoenzyme
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-12-23 Classification: HYDROLASE Ligands: MG |
 |
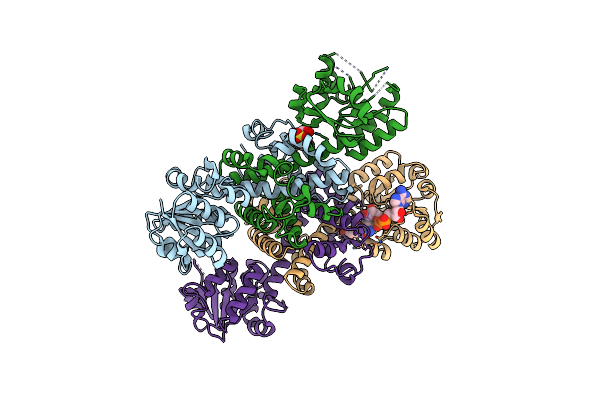
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2020-08-12 Classification: OXIDOREDUCTASE Ligands: BO3 |
 |
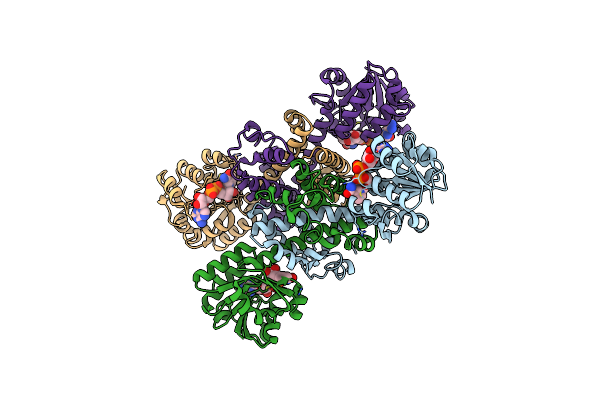
Crystal Structure Of Sla Reductase Yihu From E. Coli With Nadh And Product Dhps
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2020-08-12 Classification: OXIDOREDUCTASE Ligands: LLQ, NAD |
 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-08-12 Classification: OXIDOREDUCTASE Ligands: NAD |
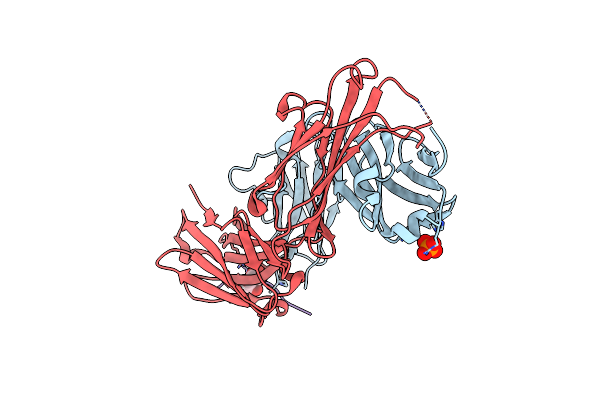 |
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-07-08 Classification: IMMUNE SYSTEM Ligands: PO4, MAN |
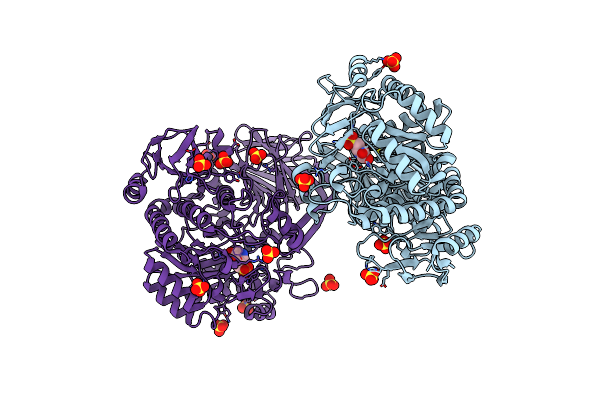 |
A Gh31 Family Sulfoquinovosidase From E. Rectale In Complex With Aza-Sugar Inhibitor Ifgsq
Organism: [eubacterium] rectale
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-07-08 Classification: Hydrolase/Hydrolas Inhibitor Ligands: 9VH, SO4 |

