Search Count: 97
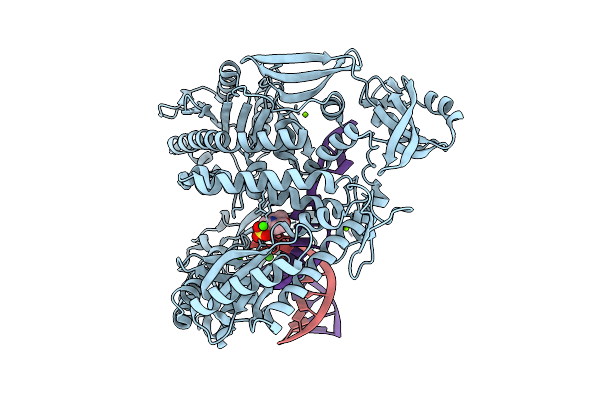 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: CA, MG, MES |
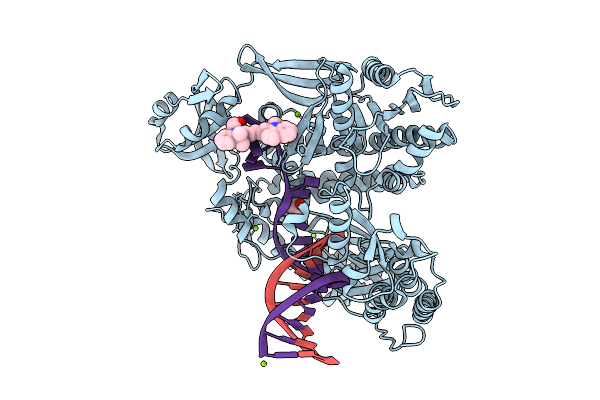 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: MG, 5CY, GOL |
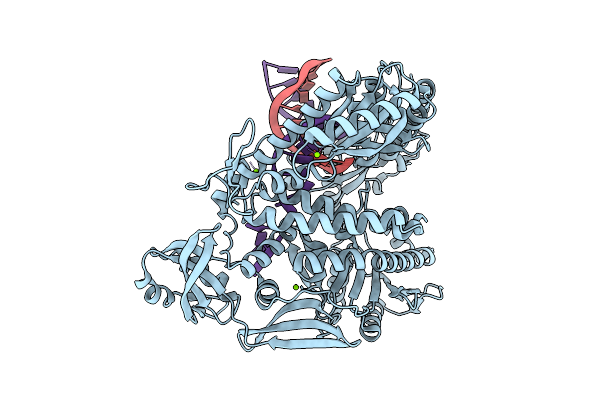 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: MG |
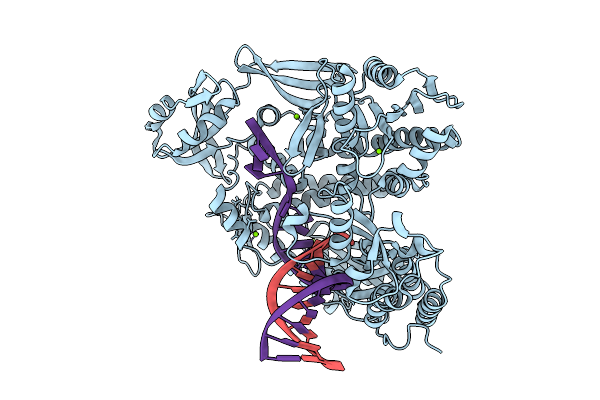 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: MG |
 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: 9O7, MG, IOD, EDO |
 |
Organism: Thermococcus kodakarensis, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-10 Classification: TRANSFERASE/DNA Ligands: 9O7, MG, IOD, CL |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: STRUCTURAL PROTEIN |
 |
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: STRUCTURAL PROTEIN |
 |
Human Pigment Epithelium-Derived Factor With Zinc Ions Crystallized In P2(1)2(1)2(1) Space Group
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: SIGNALING PROTEIN Ligands: ZN, SO4, PG0 |
 |
Human Pigment Epithelium-Derived Factor With Zinc Ion Crystallized In P22(1)2(1) Space Group
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: SIGNALING PROTEIN Ligands: ZN, PG0 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.33 Å Release Date: 2025-04-30 Classification: HYDROLASE Ligands: SO4, MG |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.04 Å Release Date: 2025-04-30 Classification: HYDROLASE Ligands: SO4, MG, A1AU6 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.42 Å Release Date: 2025-03-19 Classification: HYDROLASE Ligands: MG, SO4 |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.44 Å Release Date: 2025-03-19 Classification: HYDROLASE Ligands: SO4, MG, A1A9B |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.44 Å Release Date: 2025-03-19 Classification: HYDROLASE Ligands: SO4, A1A9B, MG |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2025-03-19 Classification: HYDROLASE Ligands: SO4, MG, A1AHA, MPD, ACY |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2025-03-19 Classification: TRANSFERASE Ligands: PLS, HZI |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: PROTEIN FIBRIL |
 |
Crystallographic Structure Of Ruminococcus Champanellensis Xylanase (Rcxyn30A)
Organism: Ruminococcus champanellensis 18p13 = jcm 17042
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2024-12-25 Classification: HYDROLASE Ligands: EDO, PEG, NA, F, PO4 |

